Integrative "omic" analysis of experimental bacteremia identifies a metabolic signature that distinguishes human sepsis from systemic inflammatory response syndromes
- PMID: 25054455
- PMCID: PMC4214130
- DOI: 10.1164/rccm.201404-0624OC
Integrative "omic" analysis of experimental bacteremia identifies a metabolic signature that distinguishes human sepsis from systemic inflammatory response syndromes
Abstract
Rationale: Sepsis is a leading cause of morbidity and mortality. Currently, early diagnosis and the progression of the disease are difficult to make. The integration of metabolomic and transcriptomic data in a primate model of sepsis may provide a novel molecular signature of clinical sepsis.
Objectives: To develop a biomarker panel to characterize sepsis in primates and ascertain its relevance to early diagnosis and progression of human sepsis.
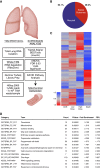
Methods: Intravenous inoculation of Macaca fascicularis with Escherichia coli produced mild to severe sepsis, lung injury, and death. Plasma samples were obtained before and after 1, 3, and 5 days of E. coli challenge and at the time of killing. At necropsy, blood, lung, kidney, and spleen samples were collected. An integrative analysis of the metabolomic and transcriptomic datasets was performed to identify a panel of sepsis biomarkers.
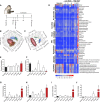
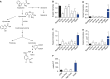
Measurements and main results: The extent of E. coli invasion, respiratory distress, lethargy, and mortality was dependent on the bacterial dose. Metabolomic and transcriptomic changes characterized severe infections and death, and indicated impaired mitochondrial, peroxisomal, and liver functions. Analysis of the pulmonary transcriptome and plasma metabolome suggested impaired fatty acid catabolism regulated by peroxisome-proliferator activated receptor signaling. A representative four-metabolite model effectively diagnosed sepsis in primates (area under the curve, 0.966) and in two human sepsis cohorts (area under the curve, 0.78 and 0.82).
Conclusions: A model of sepsis based on reciprocal metabolomic and transcriptomic data was developed in primates and validated in two human patient cohorts. It is anticipated that the identified parameters will facilitate early diagnosis and management of sepsis.
Keywords: bacteremia; metabolomics; mitochondrial dysfunction; nonhuman primates; transcriptomics.
Figures

References
-
- Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303–1310. - PubMed
-
- Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369:840–851. - PubMed
-
- Liu V, Escobar GJ, Greene JD, Soule J, Whippy A, Angus DC, Iwashyna TJ. Hospital deaths in patients with sepsis from 2 independent cohorts. JAMA. 2014;312:90–92. - PubMed
-
- Rivers E, Nguyen B, Havstad S, Ressler J, Muzzin A, Knoblich B, Peterson E, Tomlanovich M Early Goal-Directed Therapy Collaborative Group. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N Engl J Med. 2001;345:1368–1377. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

