Full-length genome analyses of two new simian immunodeficiency virus (SIV) strains from mustached monkeys (C. Cephus) in Gabon illustrate a complex evolutionary history among the SIVmus/mon/gsn lineage
- PMID: 25054885
- PMCID: PMC4113797
- DOI: 10.3390/v6072880
Full-length genome analyses of two new simian immunodeficiency virus (SIV) strains from mustached monkeys (C. Cephus) in Gabon illustrate a complex evolutionary history among the SIVmus/mon/gsn lineage
Abstract
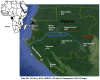
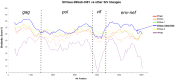
The Simian Immunodeficiency Virus (SIV) mus/mon/gsn lineage is a descendant of one of the precursor viruses to the HIV-1/SIVcpz/gor viral lineage. SIVmus and SIVgsn were sequenced from mustached and greater spot nosed monkeys in Cameroon and SIVmon from mona monkeys in Cameroon and Nigeria. In order to further document the genetic diversity of SIVmus, we analyzed two full-length genomes of new strains identified in Gabon. The whole genomes obtained showed the expected reading frames for gag, pol, vif, vpr, tat, rev, env, nef, and also for a vpu gene. Analyses showed that the Gabonese SIVmus strains were closely related and formed a monophyletic clade within the SIVmus/mon/gsn lineage. Nonetheless, within this lineage, the position of both new SIVmus differed according to the gene analyzed. In pol and nef gene, phylogenetic topologies suggested different evolutions for each of the two new SIVmus strains whereas in the other nucleic fragments studied, their positions fluctuated between SIVmon, SIVmus-1, and SIVgsn. In addition, in C1 domain of env, we identified an insertion of seven amino acids characteristic for the SIVmus/mon/gsn and HIV‑1/SIVcpz/SIVgor lineages. Our results show a high genetic diversity of SIVmus in mustached monkeys and suggest cross-species transmission events and recombination within SIVmus/mon/gsn lineage. Additionally, in Central Africa, hunters continue to be exposed to these simian viruses, and this represents a potential threat to humans.
Figures




References
-
- Beer B.E., Bailes E., Goeken R., Dapolito G., Coulibaly C., Norley S.G., Kurth R., Gautier J.P., Gautier-Hion A., Vallet D., et al. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): Evidence for host-dependent evolution of SIV within the C. Lhoesti superspecies. J. Virol. 1999;73:7734–7744. - PMC - PubMed
-
- Bibollet-Ruche F., Bailes E., Gao F., Pourrut X., Barlow K.L., Clewley J.P., Mwenda J.M., Langat D.K., Chege G.K., McClure H.M., et al. New simian immunodeficiency virus infecting de brazza’s monkeys (Cercopithecus neglectus): Evidence for a Cercopithecus monkey virus clade. J. Virol. 2004;78:7748–7762. doi: 10.1128/JVI.78.14.7748-7762.2004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

