New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae
- PMID: 25056309
- PMCID: PMC4150775
- DOI: 10.1093/nar/gku663
New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae
Abstract
Stop codon readthrough may be promoted by the nucleotide environment or drugs. In such cases, ribosomes incorporate a natural suppressor tRNA at the stop codon, leading to the continuation of translation in the same reading frame until the next stop codon and resulting in the expression of a protein with a new potential function. However, the identity of the natural suppressor tRNAs involved in stop codon readthrough remains unclear, precluding identification of the amino acids incorporated at the stop position. We established an in vivo reporter system for identifying the amino acids incorporated at the stop codon, by mass spectrometry in the yeast Saccharomyces cerevisiae. We found that glutamine, tyrosine and lysine were inserted at UAA and UAG codons, whereas tryptophan, cysteine and arginine were inserted at UGA codon. The 5' nucleotide context of the stop codon had no impact on the identity or proportion of amino acids incorporated by readthrough. We also found that two different glutamine tRNA(Gln) were used to insert glutamine at UAA and UAG codons. This work constitutes the first systematic analysis of the amino acids incorporated at stop codons, providing important new insights into the decoding rules used by the ribosome to read the genetic code.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
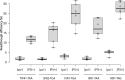
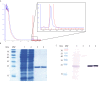


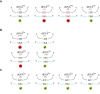
References
-
- Loh P.G., Song H. Structural and mechanistic insights into translation termination. Curr. Opin. Struct. Biol. 2010;20:98–103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

