DNA break mapping reveals topoisomerase II activity genome-wide
- PMID: 25056547
- PMCID: PMC4139894
- DOI: 10.3390/ijms150713111
DNA break mapping reveals topoisomerase II activity genome-wide
Abstract
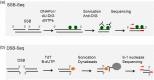
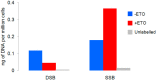
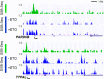
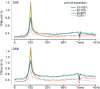
Genomic DNA is under constant assault by endogenous and exogenous DNA damaging agents. DNA breakage can represent a major threat to genome integrity but can also be necessary for genome function. Here we present approaches to map DNA double-strand breaks (DSBs) and single-strand breaks (SSBs) at the genome-wide scale by two methods called DSB- and SSB-Seq, respectively. We tested these methods in human colon cancer cells and validated the results using the Topoisomerase II (Top2)-poisoning agent etoposide (ETO). Our results show that the combination of ETO treatment with break-mapping techniques is a powerful method to elaborate the pattern of Top2 enzymatic activity across the genome.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

