Molecular characterization of a lizard adenovirus reveals the first atadenovirus with two fiber genes and the first adenovirus with either one short or three long fibers per penton
- PMID: 25056898
- PMCID: PMC4178792
- DOI: 10.1128/JVI.00306-14
Molecular characterization of a lizard adenovirus reveals the first atadenovirus with two fiber genes and the first adenovirus with either one short or three long fibers per penton
Abstract
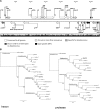
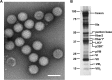
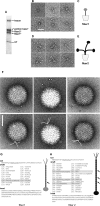
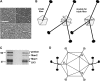
Although adenoviruses (AdVs) have been found in a wide variety of reptiles, including numerous squamate species, turtles, and crocodiles, the number of reptilian adenovirus isolates is still scarce. The only fully sequenced reptilian adenovirus, snake adenovirus 1 (SnAdV-1), belongs to the Atadenovirus genus. Recently, two new atadenoviruses were isolated from a captive Gila monster (Heloderma suspectum) and Mexican beaded lizards (Heloderma horridum). Here we report the full genomic and proteomic characterization of the latter, designated lizard adenovirus 2 (LAdV-2). The double-stranded DNA (dsDNA) genome of LAdV-2 is 32,965 bp long, with an average G+C content of 44.16%. The overall arrangement and gene content of the LAdV-2 genome were largely concordant with those in other atadenoviruses, except for four novel open reading frames (ORFs) at the right end of the genome. Phylogeny reconstructions and plesiomorphic traits shared with SnAdV-1 further supported the assignment of LAdV-2 to the Atadenovirus genus. Surprisingly, two fiber genes were found for the first time in an atadenovirus. After optimizing the production of LAdV-2 in cell culture, we determined the protein compositions of the virions. The two fiber genes produce two fiber proteins of different sizes that are incorporated into the viral particles. Interestingly, the two different fiber proteins assemble as either one short or three long fiber projections per vertex. Stoichiometry estimations indicate that the long fiber triplet is present at only one or two vertices per virion. Neither triple fibers nor a mixed number of fibers per vertex had previously been reported for adenoviruses or any other virus.
Importance: Here we show that a lizard adenovirus, LAdV-2, has a penton architecture never observed before. LAdV-2 expresses two fiber proteins-one short and one long. In the virion, most vertices have one short fiber, but a few of them have three long fibers attached to the same penton base. This observation raises new intriguing questions on virus structure. How can the triple fiber attach to a pentameric vertex? What determines the number and location of each vertex type in the icosahedral particle? Since fibers are responsible for primary attachment to the host, this novel architecture also suggests a novel mode of cell entry for LAdV-2. Adenoviruses have a recognized potential in nanobiomedicine, but only a few of the more than 200 types found so far in nature have been characterized in detail. Exploring the taxonomic wealth of adenoviruses should improve our chances to successfully use them as therapeutic tools.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- Harrach B, Benkő M, Both G, Brown M, Davison A, Echavarría M, Hess M, Jones M, Kajon A, Lehmkuhl H, Mautner V, Mittal S, Wadell G. 2011. Family Adenoviridae, p 95–111 In King A, Adams M, Carstens E, Lefkowitz E. (ed), Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier, San Diego, CA
-
- Benkő M, Doszpoly A. 2011. Ichtadenovirus. Adenoviridae, p 29–32 In Tidona CA, Darai G. (ed), The Springer index of viruses. Springer, New York, NY
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

