The Caenorhabditis elegans SUN protein UNC-84 interacts with lamin to transfer forces from the cytoplasm to the nucleoskeleton during nuclear migration
- PMID: 25057012
- PMCID: PMC4161519
- DOI: 10.1091/mbc.E14-05-0971
The Caenorhabditis elegans SUN protein UNC-84 interacts with lamin to transfer forces from the cytoplasm to the nucleoskeleton during nuclear migration
Abstract
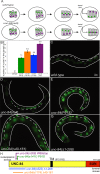
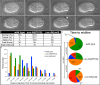
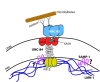
Nuclear migration is a critical component of many cellular and developmental processes. The nuclear envelope forms a barrier between the cytoplasm, where mechanical forces are generated, and the nucleoskeleton. The LINC complex consists of KASH proteins in the outer nuclear membrane and SUN proteins in the inner nuclear membrane that bridge the nuclear envelope. How forces are transferred from the LINC complex to the nucleoskeleton is poorly understood. The Caenorhabditis elegans lamin, LMN-1, is required for nuclear migration and interacts with the nucleoplasmic domain of the SUN protein UNC-84. This interaction is weakened by the unc-84(P91S) missense mutation. These mutant nuclei have an intermediate nuclear migration defect-live imaging of nuclei or LMN-1::GFP shows that many nuclei migrate normally, others initiate migration before subsequently failing, and others fail to begin migration. At least one other component of the nucleoskeleton, the NET5/Samp1/Ima1 homologue SAMP-1, plays a role in nuclear migration. We propose a nut-and-bolt model to explain how forces are dissipated across the nuclear envelope during nuclear migration. In this model, SUN/KASH bridges serve as bolts through the nuclear envelope, and nucleoskeleton components LMN-1 and SAMP-1 act as both nuts and washers on the inside of the nucleus.
© 2014 Bone et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures





References
-
- Altun ZF, Hall DH. Epithelial system, hypodermis. 2009 In: WormAtlas, DOI:10.3908/wormatlas.1.13.
-
- Borrego-Pinto J, Jegou T, Osorio DS, Aurade F, Gorjanacz M, Koch B, Mattaj IW, Gomes ER. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci. 2012;125:1099–1105. - PubMed
-
- Bruusgaard JC, Liestol K, Gundersen K. Distribution of myonuclei and microtubules in live muscle fibers of young, middle-aged, and old mice. J Appl Physiol. 2006;100:2024–2030. - PubMed
-
- Buch C, Lindberg R, Figueroa R, Gudise S, Onischenko E, Hallberg E. An integral protein of the inner nuclear membrane localizes to the mitotic spindle in mammalian cells. J Cell Sci. 2009;122:2100–2107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

