Inherited CARD9 deficiency in 2 unrelated patients with invasive Exophiala infection
- PMID: 25057046
- PMCID: PMC4447834
- DOI: 10.1093/infdis/jiu412
Inherited CARD9 deficiency in 2 unrelated patients with invasive Exophiala infection
Abstract
Background: Exophiala species are mostly responsible for skin infections. Invasive Exophiala dermatitidis disease is a rare and frequently fatal infection, with 42 cases reported. About half of these cases had no known risk factors. Similarly, invasive Exophiala spinifera disease is extremely rare, with only 3 cases reported, all in patients with no known immunodeficiency. Autosomal recessive CARD9 deficiency has recently been reported in otherwise healthy patients with severe fungal diseases caused by Candida species, dermatophytes, or Phialophora verrucosa.
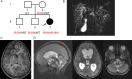
Methods: We investigated an 8-year-old girl from a nonconsanguineous Angolan kindred, who was born in France and developed disseminated E. dermatitidis disease and a 26 year-old woman from an Iranian consaguineous kindred, who was living in Iran and developed disseminated E. spinifera disease. Both patients were otherwise healthy.
Results: We sequenced CARD9 and found both patients to be homozygous for loss-of-function mutations (R18W and E323del). The first patient had segmental uniparental disomy of chromosome 9, carrying 2 copies of the maternal CARD9 mutated allele.
Conclusions: These are the first 2 patients with inherited CARD9 deficiency and invasive Exophiala disease to be described. CARD9 deficiency should thus be considered in patients with unexplained invasive Exophiala species disease, even in the absence of other infections.
Keywords: Exophiala species; autosomal recessive CARD9 deficiency; central nervous system; invasive fungal infection; osteomyelitis; parental unidisomy.
© The Author 2014. Published by Oxford University Press on behalf of the Infectious Diseases Society of America. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures



Comment in
-
Human invasive mycoses: immunogenetics on the rise.J Infect Dis. 2015 Apr 15;211(8):1205-7. doi: 10.1093/infdis/jiu411. Epub 2014 Jul 23. J Infect Dis. 2015. PMID: 25057047 Free PMC article. No abstract available.
References
-
- Horre R, Schaal KP, Siekmeier R, Sterzik B, de Hoog GS, Schnitzler N. Isolation of fungi, especially Exophiala dermatitidis, in patients suffering from cystic fibrosis. A prospective study. Respiration 2004; 71:360–6. - PubMed
-
- de Hoog GS, Matos T, Sudhadham M, Luijsterburg KF, Haase G. Intestinal prevalence of the neurotropic black yeast Exophiala (Wangiella) dermatitidis in healthy and impaired individuals. Mycoses 2005; 48:142–5. - PubMed
-
- Patel AK, Patel KK, Darji P, Singh R, Shivaprakash MR, Chakrabarti A. Exophiala dermatitidis endocarditis on native aortic valve in a postrenal transplant patient and review of literature on E. dermatitidis infections. Mycoses 2013; 56:365–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

