Mutant huntingtin affects cortical progenitor cell division and development of the mouse neocortex
- PMID: 25057205
- PMCID: PMC6608303
- DOI: 10.1523/JNEUROSCI.0715-14.2014
Mutant huntingtin affects cortical progenitor cell division and development of the mouse neocortex
Abstract
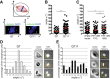
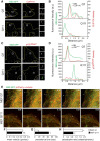
A polyglutamine expansion in huntingtin (HTT) causes the specific death of adult neurons in Huntington's disease (HD). Most studies have thus focused on mutant HTT (mHTT) toxicity in adulthood, and its developmental effects have been largely overlooked. We found that mHTT caused mitotic spindle misorientation in cultured cells by altering the localization of dynein, NuMA, and the p150(Glued) subunit of dynactin to the spindle pole and cell cortex and of CLIP170 and p150(Glued) to microtubule plus-ends. mHTT also affected spindle orientation in dividing mouse cortical progenitors, altering the thickness of the developing cortex. The serine/threonine kinase Akt, which regulates HTT function, rescued the spindle misorientation caused by the mHTT, by serine 421 (S421) phosphorylation, in cultured cells and in mice. Thus, cortical development is affected in HD, and this early defect can be rescued by HTT phosphorylation at S421.
Keywords: Huntington disease; cortical neurogenesis; mitosis; spindle orientation.
Copyright © 2014 the authors 0270-6474/14/3410034-07$15.00/0.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
