Silencing is noisy: population and cell level noise in telomere-adjacent genes is dependent on telomere position and sir2
- PMID: 25057900
- PMCID: PMC4109849
- DOI: 10.1371/journal.pgen.1004436
Silencing is noisy: population and cell level noise in telomere-adjacent genes is dependent on telomere position and sir2
Abstract
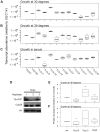
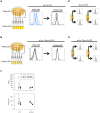
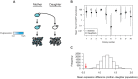
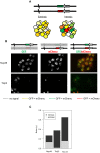
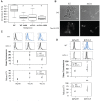
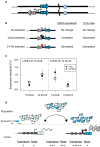
Cell-to-cell gene expression noise is thought to be an important mechanism for generating phenotypic diversity. Furthermore, telomeric regions are major sites for gene amplification, which is thought to drive genetic diversity. Here we found that individual subtelomeric TLO genes exhibit increased variation in transcript and protein levels at both the cell-to-cell level as well as at the population-level. The cell-to-cell variation, termed Telomere-Adjacent Gene Expression Noise (TAGEN) was largely intrinsic noise and was dependent upon genome position: noise was reduced when a TLO gene was expressed at an ectopic internal locus and noise was elevated when a non-telomeric gene was expressed at a telomere-adjacent locus. This position-dependent TAGEN also was dependent on Sir2p, an NAD+-dependent histone deacetylase. Finally, we found that telomere silencing and TAGEN are tightly linked and regulated in cis: selection for either silencing or activation of a TLO-adjacent URA3 gene resulted in reduced noise at the neighboring TLO but not at other TLO genes. This provides experimental support to computational predictions that the ability to shift between silent and active chromatin states has a major effect on cell-to-cell noise. Furthermore, it demonstrates that these shifts affect the degree of expression variation at each telomere individually.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, et al. (2006) Noise in protein expression scales with natural protein abundance. Nat Genet 38: 636–643. - PubMed
-
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, et al. (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846. - PubMed
-
- Hernday AD, Braaten BA, Broitman-Maduro G, Engelberts P, Low DA (2004) Regulation of the pap epigenetic switch by CpxAR: phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol Cell 16: 537–547. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

