8.2% of the Human genome is constrained: variation in rates of turnover across functional element classes in the human lineage
- PMID: 25057982
- PMCID: PMC4109858
- DOI: 10.1371/journal.pgen.1004525
8.2% of the Human genome is constrained: variation in rates of turnover across functional element classes in the human lineage
Abstract
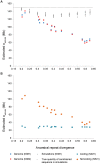
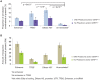
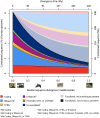
Ten years on from the finishing of the human reference genome sequence, it remains unclear what fraction of the human genome confers function, where this sequence resides, and how much is shared with other mammalian species. When addressing these questions, functional sequence has often been equated with pan-mammalian conserved sequence. However, functional elements that are short-lived, including those contributing to species-specific biology, will not leave a footprint of long-lasting negative selection. Here, we address these issues by identifying and characterising sequence that has been constrained with respect to insertions and deletions for pairs of eutherian genomes over a range of divergences. Within noncoding sequence, we find increasing amounts of mutually constrained sequence as species pairs become more closely related, indicating that noncoding constrained sequence turns over rapidly. We estimate that half of present-day noncoding constrained sequence has been gained or lost in approximately the last 130 million years (half-life in units of divergence time, d1/2 = 0.25-0.31). While enriched with ENCODE biochemical annotations, much of the short-lived constrained sequences we identify are not detected by models optimized for wider pan-mammalian conservation. Constrained DNase 1 hypersensitivity sites, promoters and untranslated regions have been more evolutionarily stable than long noncoding RNA loci which have turned over especially rapidly. By contrast, protein coding sequence has been highly stable, with an estimated half-life of over a billion years (d1/2 = 2.1-5.0). From extrapolations we estimate that 8.2% (7.1-9.2%) of the human genome is presently subject to negative selection and thus is likely to be functional, while only 2.2% has maintained constraint in both human and mouse since these species diverged. These results reveal that the evolutionary history of the human genome has been highly dynamic, particularly for its noncoding yet biologically functional fraction.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Pennisi E (2012) Genomics. ENCODE project writes eulogy for junk DNA. Science 337: 1159, 1161. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

