Protein profiling of human lung telocytes and microvascular endothelial cells using iTRAQ quantitative proteomics
- PMID: 25059386
- PMCID: PMC4508144
- DOI: 10.1111/jcmm.12350
Protein profiling of human lung telocytes and microvascular endothelial cells using iTRAQ quantitative proteomics
Abstract
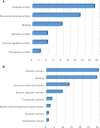
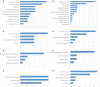
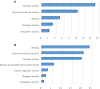
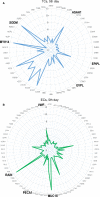
Telocytes (TCs) are described as a particular type of cells of the interstitial space (www.telocytes.com). Their main characteristics are the very long telopodes with alternating podoms and podomers. Recently, we performed a comparative proteomic analysis of human lung TCs with fibroblasts, demonstrating that TCs are clearly a distinct cell type. Therefore, the present study aims to reinforce this idea by comparing lung TCs with endothelial cells (ECs), since TCs and ECs share immunopositivity for CD34. We applied isobaric tag for relative and absolute quantification (iTRAQ) combined with automated 2-D nano-ESI LC-MS/MS to analyse proteins extracted from TCs and ECs in primary cell cultures. In total, 1609 proteins were identified in cell cultures. 98 proteins (the 5th day), and 82 proteins (10th day) were confidently quantified (screened by two-sample t-test, P < 0.05) as up- or down-regulated (fold change >2). We found that in TCs there are 38 up-regulated proteins at the 5th day and 26 up-regulated proteins at the 10th day. Bioinformatics analysis using Panther revealed that the 38 proteins associated with TCs represented cellular functions such as intercellular communication (via vesicle mediated transport) and structure morphogenesis, being mainly cytoskeletal proteins and oxidoreductases. In addition, we found 60 up-regulated proteins in ECs e.g.: cell surface glycoprotein MUC18 (15.54-fold) and von Willebrand factor (5.74-fold). The 26 up-regulated proteins in TCs at 10th day, were also analysed and confirmed the same major cellular functions, while the 56 down-regulated proteins confirmed again their specificity for ECs. In conclusion, we report here the first extensive comparison of proteins from TCs and ECs using a quantitative proteomics approach. Our data show that TCs are completely different from ECs. Protein expression profile showed that TCs play specific roles in intercellular communication and intercellular signalling. Moreover, they might inhibit the oxidative stress and cellular ageing and may have pro-proliferative effects through the inhibition of apoptosis. The group of proteins identified in this study needs to be explored further for the role in pathogenesis of lung disease.
Keywords: LC-MS/MS; iTRAQ; intercellular signalling; lung; microvascular endothelial cells; proteomics; telocytes.
© 2014 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

