Inhibition and mechanism of HDAC8 revisited
- PMID: 25060069
- PMCID: PMC4140456
- DOI: 10.1021/ja501548p
Inhibition and mechanism of HDAC8 revisited
Abstract
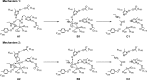
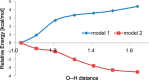
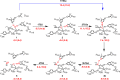
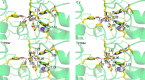
Histone deacetylases (HDACs) have found intense interest as drug targets for a variety of diseases, but there is disagreement about basic aspects of the inhibition and mechanism of HDACs. QM/MM calculations of HDAC8 including a large QM region provide a model that is consistent with the available crystal structures and structure-activity relationships of different HDAC inhibitors. The calculations support a spontaneous proton transfer from a hydroxamic acid to an active site histidine upon binding to the zinc. The role of the H142/D176 catalytic dyad as the general base of the reaction is elucidated. The reasons for the disagreements between previous proposals are discussed. The results provide detailed insights into the unique mechanism of HDACs, including the role of the two catalytic dyads and function of the potassium near the active site. They also have important implications for the design of novel inhibitors for a number of HDACs such as the class IIa HDACs.
Figures


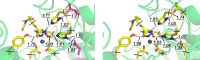
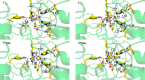
References
-
- Arrowsmith C. H.; Bountra C.; Fish P. V.; Lee K.; Schapira M. Nat. Rev. Drug Discovery 2012, 11, 384. - PubMed
-
- Dawson M. A.; Kouzarides T. Cell 2012, 150, 12. - PubMed
-
- Lane A. A.; Chabner B. A. J. Clin. Oncol. 2009, 27, 5459. - PubMed
-
- Carew J. S.; Giles F. J.; Nawrocki S. T. Cancer Lett. 2008, 269, 7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

