Thermodynamics of Deca-alanine Folding in Water
- PMID: 25061447
- PMCID: PMC4095909
- DOI: 10.1021/ct5002076
Thermodynamics of Deca-alanine Folding in Water
Abstract
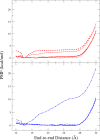
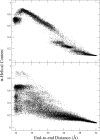
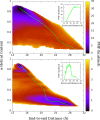
The determination of the folding dynamics of polypeptides and proteins is critical in characterizing their functions in biological systems. Numerous computational models and methods have been developed for studying structure formation at the atomic level. Due to its small size and simple structure, deca-alanine is used as a model system in molecular dynamics (MD) simulations. The free energy of unfolding in vacuum has been studied extensively using the end-to-end distance of the peptide as the reaction coordinate. However, few studies have been conducted in the presence of explicit solvent. Previous results show a significant decrease in the free energy of extended conformations in water, but the α-helical state is still notably favored over the extended state. Although sufficient in vacuum, we show that end-to-end distance is incapable of capturing the full complexity of deca-alanine folding in water. Using α-helical content as a second reaction coordinate, we deduce a more descriptive free-energy landscape, one which reveals a second energy minimum in the extended conformations that is of comparable free energy to the α-helical state. Equilibrium simulations demonstrate the relative stability of the extended and α-helical states in water as well as the transition between the two states. This work reveals both the necessity and challenge of determining a proper reaction coordinate to fully characterize a given process.
Figures





Similar articles
-
Thermodynamics and mechanism of alpha helix initiation in alanine and valine peptides.Biochemistry. 1991 Jun 18;30(24):6059-70. doi: 10.1021/bi00238a033. Biochemistry. 1991. PMID: 2043644
-
Conformational equilibrium in alanine-rich peptides probed by reversible stretching simulations.J Phys Chem B. 2006 Aug 24;110(33):16718-23. doi: 10.1021/jp0601116. J Phys Chem B. 2006. PMID: 16913811
-
Simulation of coupled folding and binding of an intrinsically disordered protein in explicit solvent with metadynamics.J Mol Graph Model. 2016 Jul;68:114-127. doi: 10.1016/j.jmgm.2016.06.015. Epub 2016 Jul 1. J Mol Graph Model. 2016. PMID: 27423742
-
Long timestep dynamics of peptides by the dynamics driver approach.Proteins. 1995 Apr;21(4):282-302. doi: 10.1002/prot.340210403. Proteins. 1995. PMID: 7567951
-
Protein and peptide folding explored with molecular simulations.Acc Chem Res. 2002 Jun;35(6):447-54. doi: 10.1021/ar0100172. Acc Chem Res. 2002. PMID: 12069630 Review.
Cited by
-
Peptide Dynamics and Metadynamics: Leveraging Enhanced Sampling Molecular Dynamics to Robustly Model Long-Timescale Transitions.Methods Mol Biol. 2022;2405:151-167. doi: 10.1007/978-1-0716-1855-4_8. Methods Mol Biol. 2022. PMID: 35298813 Free PMC article.
-
Thermodynamics of helix formation in small peptides of varying length in vacuo, in implicit solvent, and in explicit solvent.J Mol Model. 2018 Dec 12;25(1):3. doi: 10.1007/s00894-018-3886-2. J Mol Model. 2018. PMID: 30542771
-
Integrating solvation shell structure in experimentally driven molecular dynamics using x-ray solution scattering data.J Chem Phys. 2020 May 29;152(20):204115. doi: 10.1063/5.0007158. J Chem Phys. 2020. PMID: 32486681 Free PMC article.
-
Molecular Mechanism of Processive 3' to 5' RNA Translocation in the Active Subunit of the RNA Exosome Complex.J Am Chem Soc. 2016 Mar 30;138(12):4069-78. doi: 10.1021/jacs.5b12065. Epub 2016 Mar 21. J Am Chem Soc. 2016. PMID: 26928279 Free PMC article.
-
Statistical Mechanical Design Principles for Coarse-Grained Interactions across Different Conformational Free Energy Surfaces.J Phys Chem Lett. 2023 Feb 16;14(6):1354-1362. doi: 10.1021/acs.jpclett.2c03844. Epub 2023 Feb 2. J Phys Chem Lett. 2023. PMID: 36728761 Free PMC article.
References
-
- Anfinsen C. B. Principles that govern the folding of protein chains. Science 1973, 181, 223–227. - PubMed
-
- Baldwin R. L.; Rose G. D. Is protein folding hierarchic? II. Folding intermediates and transition states. Trends Biochem. Sci. 1999, 24, 77–83. - PubMed
-
- Baldwin R. L.; Rose G. D. Is protein folding hierarchic? I. Local structure and peptide folding. Trends Biochem. Sci. 1999, 24, 26–33. - PubMed
-
- Williams S.; Causgrove T. P.; Gilmanshin R.; Fang K. S.; Callender R. H.; Woodruff W. H.; Dyer R. B. Fast events in protein folding: Helix melting and formation in a small peptide. Biochemistry 1996, 35, 691–697. - PubMed
-
- Young W. S.; Brooks C. L. III. A microscopic view of helix propagation: N and C-terminal helix growth in alanine helices. J. Mol. Biol. 1996, 259, 560–572. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
