The SKIV2L RNA exosome limits activation of the RIG-I-like receptors
- PMID: 25064072
- PMCID: PMC4139417
- DOI: 10.1038/ni.2948
The SKIV2L RNA exosome limits activation of the RIG-I-like receptors
Abstract
Sensors of the innate immune system that detect intracellular nucleic acids must be regulated to prevent inappropriate activation by endogenous DNA and RNA. The exonuclease Trex1 regulates the DNA-sensing pathway by metabolizing potential DNA ligands that trigger it. However, an analogous mechanism for regulating the RIG-I-like receptors (RLRs) that detect RNA remains unknown. We found here that the SKIV2L RNA exosome potently limited the activation of RLRs. The unfolded protein response (UPR), which generated endogenous RLR ligands through the cleavage of cellular RNA by the endonuclease IRE-1, triggered the production of type I interferons in cells depleted of SKIV2L. Humans with deficiency in SKIV2L had a type I interferon signature in their peripheral blood. Our findings reveal a mechanism for the intracellular metabolism of immunostimulatory RNA, with implications for specific autoimmune disorders.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

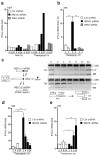
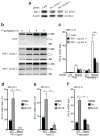
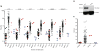
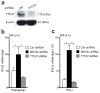
Comment in
-
RNA exosomes keep endogenous RNA under the radar.Nat Immunol. 2014 Sep;15(9):830-1. doi: 10.1038/ni.2966. Nat Immunol. 2014. PMID: 25137466 No abstract available.
References
-
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. - PubMed
-
- Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. - PubMed
-
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

