CCBuilder: an interactive web-based tool for building, designing and assessing coiled-coil protein assemblies
- PMID: 25064570
- PMCID: PMC4201159
- DOI: 10.1093/bioinformatics/btu502
CCBuilder: an interactive web-based tool for building, designing and assessing coiled-coil protein assemblies
Abstract
Motivation: The ability to accurately model protein structures at the atomistic level underpins efforts to understand protein folding, to engineer natural proteins predictably and to design proteins de novo. Homology-based methods are well established and produce impressive results. However, these are limited to structures presented by and resolved for natural proteins. Addressing this problem more widely and deriving truly ab initio models requires mathematical descriptions for protein folds; the means to decorate these with natural, engineered or de novo sequences; and methods to score the resulting models.
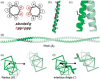
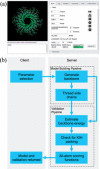
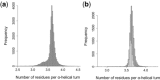
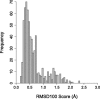
Results: We present CCBuilder, a web-based application that tackles the problem for a defined but large class of protein structure, the α-helical coiled coils. CCBuilder generates coiled-coil backbones, builds side chains onto these frameworks and provides a range of metrics to measure the quality of the models. Its straightforward graphical user interface provides broad functionality that allows users to build and assess models, in which helix geometry, coiled-coil architecture and topology and protein sequence can be varied rapidly. We demonstrate the utility of CCBuilder by assembling models for 653 coiled-coil structures from the PDB, which cover >96% of the known coiled-coil types, and by generating models for rarer and de novo coiled-coil structures.
Availability and implementation: CCBuilder is freely available, without registration, at http://coiledcoils.chm.bris.ac.uk/app/cc_builder/.
© The Author 2014. Published by Oxford University Press.
Figures

References
-
- Bansal M, et al. HELANAL: a program to characterize helix geometry in proteins. J. Biomol. Struct. Dyn. 2000;17:811–819. - PubMed
-
- Bowie JU, et al. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. - PubMed
-
- Bradley P, et al. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–1871. - PubMed
-
- Burton AJ, et al. Accessibility, reactivity, and selectivity of side chains within a channel of de novo peptide assembly. J. Am. Chem. Soc. 2013;135:12524–12527. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

