Screening and identification of small molecule inhibitors of ErbB2-induced invasion
- PMID: 25070180
- PMCID: PMC5528609
- DOI: 10.1016/j.molonc.2014.07.004
Screening and identification of small molecule inhibitors of ErbB2-induced invasion
Abstract
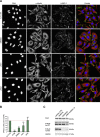
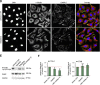
ERBB2 amplification and overexpression are strongly associated with invasive cancer with high recurrence and poor prognosis. Enhanced ErbB2 signaling induces cysteine cathepsin B and L expression leading to their higher proteolytic activity (zFRase activity), which is crucial for the invasion of ErbB2-positive breast cancer cells in vitro. Here we introduce a simple screening system based on zFRase activity as a primary readout and a following robust invasion assay and lysosomal distribution analysis for the identification of compounds that can inhibit ErbB2-induced invasion. With an unbiased kinase inhibitor screen, we identified Bohemine/Roscovitine, Gö6979 and JAK3 inhibitor VI as compounds that can efficiently decrease cysteine cathepsin activity. Using the well-established and clinically relevant ErbB1 and ErbB2 inhibitor lapatinib as a positive control, we studied their ability to inhibit ErbB2-induced invasion in 3-dimensional Matrigel cultures. We found one of them, JAK3 inhibitor VI, capable of inhibiting invasion of highly invasive ErbB2-positive ovarian cancer cells as efficiently as lapatinib, whereas Gö6979 and Roscovitine displayed more modest inhibition. All compounds reversed the malignant, ErbB2-induced and invasion-supporting peripheral distribution of lysosomes. This effect was most evident for lapatinib and JAK3 inhibitor VI and milder for Gö6979 and Roscovitine. Our results further showed that JAK3 inhibitor VI function was independent of JAK kinases but involved downregulation of cathepsin L. We postulate that the screening method and the verification experiments that are based on oncogene-induced changes in lysosomal hydrolase activity and lysosomal distribution could be used for identification of novel inhibitors of ErbB2-induced invasiveness. Additionally, we introduce a novel function for lapatinib in controlling malignant lysosomal distribution, that may also be involved in its capability to inhibit ErbB2-induced invasion in vivo.
Keywords: JAK3 inhibitor VI; Lapatinib; Lysosome; Ovarian cancer; Three-dimensional invasion assay; cathepsin L.
Copyright © 2014 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
References
-
- Adams, C. , Aldous, D.J. , Amendola, S. , Bamborough, P. , Bright, C. , Crowe, S. , Eastwood, P. , Fenton, G. , Foster, M. , Harrison, T.K. , King, S. , Lai, J. , Lawrence, C. , Letallec, J.P. , McCarthy, C. , Moorcroft, N. , Page, K. , Rao, S. , Redford, J. , Sadiq, S. , Smith, K. , Souness, J.E. , Thurairatnam, S. , Vine, M. , Wyman, B. , 2003. Mapping the kinase domain of Janus Kinase 3. Bioorg. Med. Chem. Lett. 13, 3105–3110. - PubMed
-
- Arribas, J. , Baselga, J. , Pedersen, K. , Parra-Palau, J.L. , 2011. p95HER2 and breast cancer. Cancer Res. 71, 1515–1519. - PubMed
-
- Baselga, J. , Swain, S.M. , 2009. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat. Rev. Cancer 9, 463–475. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

