Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen
- PMID: 25070509
- PMCID: PMC4231636
- DOI: 10.1093/gbe/evu160
Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen
Abstract
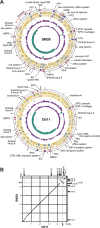
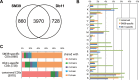
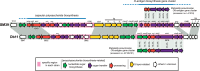
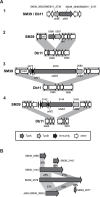
Serratia marcescens is an important nosocomial pathogen that can cause an array of infections, most notably of the urinary tract and bloodstream. Naturally, it is found in many environmental niches, and is capable of infecting plants and animals. The emergence and spread of multidrug-resistant strains producing extended-spectrum or metallo beta-lactamases now pose a threat to public health worldwide. Here we report the complete genome sequences of two carefully selected S. marcescens strains, a multidrug-resistant clinical isolate (strain SM39) and an insect isolate (strain Db11). Our comparative analyses reveal the core genome of S. marcescens and define the potential metabolic capacity, virulence, and multidrug resistance of this species. We show a remarkable intraspecies genetic diversity, both at the sequence level and with regards genome flexibility, which may reflect the diversity of niches inhabited by members of this species. A broader analysis with other Serratia species identifies a set of approximately 3,000 genes that characterize the genus. Within this apparent genetic diversity, we identified many genes implicated in the high virulence potential and antibiotic resistance of SM39, including the metallo beta-lactamase and multiple other drug resistance determinants carried on plasmid pSMC1. We further show that pSMC1 is most closely related to plasmids circulating in Pseudomonas species. Our data will provide a valuable basis for future studies on S. marcescens and new insights into the genetic mechanisms that underlie the emergence of pathogens highly resistant to multiple antimicrobial agents.
Keywords: Serratia marcescens; genome plasticity; multidrug resistance; virulence.
© The Author(s) 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. - PubMed
-
- Aucken HM, Pitt TL. Serological relationships of the O antigens of Klebsiella pneumoniae O5, Escherichia coli O8 and a new O serotype of Serratia marcescens. FEMS Microbiol Lett. 1991;64:93–97. - PubMed
-
- Chen J, Kuroda T, Huda MN, Mizushima T, Tsuchiya T. An RND-type multidrug efflux pump SdeXY from Serratia marcescens. J Antimicrob Chemother. 2003;52:176–179. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

