Bacillus subtilis chromosome organization oscillates between two distinct patterns
- PMID: 25071173
- PMCID: PMC4156703
- DOI: 10.1073/pnas.1407461111
Bacillus subtilis chromosome organization oscillates between two distinct patterns
Abstract
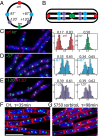
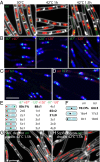
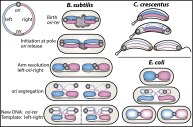
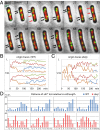
Bacterial chromosomes have been found to possess one of two distinct patterns of spatial organization. In the first, called "ori-ter" and exemplified by Caulobacter crescentus, the chromosome arms lie side-by-side, with the replication origin and terminus at opposite cell poles. In the second, observed in slow-growing Escherichia coli ("left-ori-right"), the two chromosome arms reside in separate cell halves, on either side of a centrally located origin. These two patterns, rotated 90° relative to each other, appear to result from different segregation mechanisms. Here, we show that the Bacillus subtilis chromosome alternates between them. For most of the cell cycle, newly replicated origins are maintained at opposite poles with chromosome arms adjacent to each other, in an ori-ter configuration. Shortly after replication initiation, the duplicated origins move as a unit to midcell and the two unreplicated arms resolve into opposite cell halves, generating a left-ori-right pattern. The origins are then actively segregated toward opposite poles, resetting the cycle. Our data suggest that the condensin complex and the parABS partitioning system are the principal driving forces underlying this oscillatory cycle. We propose that the distinct organization patterns observed for bacterial chromosomes reflect a common organization-segregation mechanism, and that simple modifications to it underlie the unique patterns observed in different species.
Keywords: DNA replication; ParA; SMC condensin; chromosome segregation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Switching modes of chromosome dynamics in the bacterial cell cycle.Proc Natl Acad Sci U S A. 2014 Sep 2;111(35):12580-1. doi: 10.1073/pnas.1413657111. Epub 2014 Aug 25. Proc Natl Acad Sci U S A. 2014. PMID: 25157151 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

