Tautomerism provides a molecular explanation for the mutagenic properties of the anti-HIV nucleoside 5-aza-5,6-dihydro-2'-deoxycytidine
- PMID: 25071207
- PMCID: PMC4136561
- DOI: 10.1073/pnas.1405635111
Tautomerism provides a molecular explanation for the mutagenic properties of the anti-HIV nucleoside 5-aza-5,6-dihydro-2'-deoxycytidine
Abstract
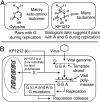
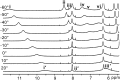
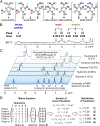
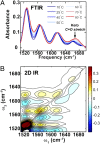
Viral lethal mutagenesis is a strategy whereby the innate immune system or mutagenic pool nucleotides increase the error rate of viral replication above the error catastrophe limit. Lethal mutagenesis has been proposed as a mechanism for several antiviral compounds, including the drug candidate 5-aza-5,6-dihydro-2'-deoxycytidine (KP1212), which causes A-to-G and G-to-A mutations in the HIV genome, both in tissue culture and in HIV positive patients undergoing KP1212 monotherapy. This work explored the molecular mechanism(s) underlying the mutagenicity of KP1212, and specifically whether tautomerism, a previously proposed hypothesis, could explain the biological consequences of this nucleoside analog. Establishing tautomerism of nucleic acid bases under physiological conditions has been challenging because of the lack of sensitive methods. This study investigated tautomerism using an array of spectroscopic, theoretical, and chemical biology approaches. Variable temperature NMR and 2D infrared spectroscopic methods demonstrated that KP1212 existed as a broad ensemble of interconverting tautomers, among which enolic forms dominated. The mutagenic properties of KP1212 were determined empirically by in vitro and in vivo replication of a single-stranded vector containing a single KP1212. It was found that KP1212 paired with both A (10%) and G (90%), which is in accord with clinical observations. Moreover, this mutation frequency is sufficient for pushing a viral population over its error catastrophe limit, as observed before in cell culture studies. Finally, a model is proposed that correlates the mutagenicity of KP1212 with its tautomeric distribution in solution.
Keywords: KP1461; site-specific mutagenesis; spectral deconvolution; viral decay acceleration; viral extinction.
Conflict of interest statement
Conflict of interest statement: J.M.E. declares a competing financial interest as a cofounder and advisor for a pharmaceutical company interested in developing mutagenic inhibitors of HIV.
Figures

Similar articles
-
Two-dimensional IR spectroscopy of the anti-HIV agent KP1212 reveals protonated and neutral tautomers that influence pH-dependent mutagenicity.Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):3229-34. doi: 10.1073/pnas.1415974112. Epub 2015 Mar 2. Proc Natl Acad Sci U S A. 2015. PMID: 25733867 Free PMC article.
-
Evaluation of anti-HIV-1 mutagenic nucleoside analogues.J Biol Chem. 2015 Jan 2;290(1):371-83. doi: 10.1074/jbc.M114.616383. Epub 2014 Nov 14. J Biol Chem. 2015. PMID: 25398876 Free PMC article.
-
5,6-Dihydro-5-aza-2'-deoxycytidine potentiates the anti-HIV-1 activity of ribonucleotide reductase inhibitors.Bioorg Med Chem. 2013 Nov 15;21(22):7222-8. doi: 10.1016/j.bmc.2013.08.023. Epub 2013 Aug 20. Bioorg Med Chem. 2013. PMID: 24120088 Free PMC article.
-
Structural Insights Into Tautomeric Dynamics in Nucleic Acids and in Antiviral Nucleoside Analogs.Front Mol Biosci. 2022 Jan 25;8:823253. doi: 10.3389/fmolb.2021.823253. eCollection 2021. Front Mol Biosci. 2022. PMID: 35145998 Free PMC article. Review.
-
Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity.Viruses. 2022 Apr 18;14(4):841. doi: 10.3390/v14040841. Viruses. 2022. PMID: 35458571 Free PMC article. Review.
Cited by
-
Environmental Effects on Guanine-Thymine Mispair Tautomerization Explored with Quantum Mechanical/Molecular Mechanical Free Energy Simulations.J Am Chem Soc. 2020 Jun 24;142(25):11183-11191. doi: 10.1021/jacs.0c03774. Epub 2020 Jun 11. J Am Chem Soc. 2020. PMID: 32459476 Free PMC article.
-
Two-dimensional IR spectroscopy of the anti-HIV agent KP1212 reveals protonated and neutral tautomers that influence pH-dependent mutagenicity.Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):3229-34. doi: 10.1073/pnas.1415974112. Epub 2015 Mar 2. Proc Natl Acad Sci U S A. 2015. PMID: 25733867 Free PMC article.
-
Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes.Nature. 2015 Mar 19;519(7543):315-20. doi: 10.1038/nature14227. Epub 2015 Mar 11. Nature. 2015. PMID: 25762137 Free PMC article.
-
Hyperpolarizing DNA Nucleobases via NMR Signal Amplification by Reversible Exchange.Molecules. 2023 Jan 25;28(3):1198. doi: 10.3390/molecules28031198. Molecules. 2023. PMID: 36770865 Free PMC article.
-
A dielectric and spectrophotometric study of the tautomerization of 2-hydroxypyridine and 2-mercaptopyridine in water.RSC Adv. 2020 Jan 13;10(4):2389-2395. doi: 10.1039/c9ra08392h. eCollection 2020 Jan 8. RSC Adv. 2020. PMID: 35494609 Free PMC article.
References
-
- Mullins JI, Jensen MA. Evolutionary dynamics of HIV-1 and the control of AIDS. Curr Top Microbiol Immunol. 2006;299:171–192. - PubMed
-
- Johnston R. HIV cure: Controversy, consensus, and a consortium. AIDS Res Hum Retroviruses. 2010;26(9):943–946. - PubMed
-
- Esté JA, Cihlar T. Current status and challenges of antiretroviral research and therapy. Antiviral Res. 2010;85(1):25–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

