Characterization and expression analysis of chymotrypsin after bacterial challenge in the mud crab, Scylla paramamosain
- PMID: 25071403
- PMCID: PMC4094611
- DOI: 10.1590/s1415-47572014005000007
Characterization and expression analysis of chymotrypsin after bacterial challenge in the mud crab, Scylla paramamosain
Abstract
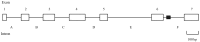
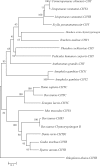
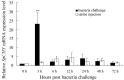
Chymotrypsin is one of the serine proteases families that have various biological functions. A chymotrypsin gene was isolated from hepatopancreas of the mud crab, Scylla paramamosain (designated SpCHY) in this study. The full-length cDNA of SpCHY contained 942 nucleotides with a polyadenylation sequence and encoded a peptide of 270 amino acids with a signal peptide of 17 amino acids. The SpCHY gene contains seven exons, six introns, a TATA box and several transcription factor binding sites that were found in 5'-promoter region which is 1221 bp in length. Real-time quantitative PCR analysis indicated that the expression level of SpCHY mRNA in hepatopancreas was significantly higher than that in other tissues. Immunocytochemistry and in situ hybridization exhibited the CHY-like reactivity presented in resorptive cells of the hepatopancreas. After bacterial challenge with Vibrio alginolyticus, the expression level of SpCHY mRNA was extremely up-regulated at 3 h in hepatopancreas. Our results suggest that SpCHY might play an important role in the mud crab's immune response.
Keywords: Scylla paramamosain; chymotrypsin; immune response; immunocytochemistry; in situ hybridization.
Figures



Similar articles
-
Identification and characterization of pro-interleukin-16 from mud crab Scylla paramamosain: The first evidence of proinflammatory cytokine in crab species.Fish Shellfish Immunol. 2017 Nov;70:701-709. doi: 10.1016/j.fsi.2017.09.057. Epub 2017 Sep 23. Fish Shellfish Immunol. 2017. PMID: 28951219
-
A clip domain serine protease regulates the expression of proPO and hemolymph clotting in mud crab, Scylla paramamosain.Fish Shellfish Immunol. 2018 Aug;79:52-64. doi: 10.1016/j.fsi.2018.05.013. Epub 2018 May 7. Fish Shellfish Immunol. 2018. PMID: 29747010
-
C-type lectin B (SpCTL-B) regulates the expression of antimicrobial peptides and promotes phagocytosis in mud crab Scylla paramamosain.Dev Comp Immunol. 2018 Jul;84:213-229. doi: 10.1016/j.dci.2018.02.016. Epub 2018 Feb 21. Dev Comp Immunol. 2018. PMID: 29476770
-
Identification and characterization of atypical 2-cysteine peroxiredoxins from mud crab Scylla paramamosain: The first evidence of two peroxiredoxin 5 genes in non-primate species and their involvement in immune defense against pathogen infection.Fish Shellfish Immunol. 2017 Oct;69:119-127. doi: 10.1016/j.fsi.2017.07.040. Epub 2017 Jul 22. Fish Shellfish Immunol. 2017. PMID: 28743622
-
SpToll1 and SpToll2 modulate the expression of antimicrobial peptides in Scylla paramamosain.Dev Comp Immunol. 2018 Oct;87:124-136. doi: 10.1016/j.dci.2018.06.008. Epub 2018 Jun 20. Dev Comp Immunol. 2018. PMID: 29935285
References
-
- Amparyup P, Jitvaropas R, Pulsook N, Tassanakajon A. Molecular cloning, characterization and expression of a masquerade-like serine proteinase homologue from black tiger shrimp Penaeus monodon. Fish Shellfish Immunol. 2007;22:535–546. - PubMed
-
- Aladaileh S, Rodney P, Nair SV, Raftos DA. Characterization of phenoloxidase activity in Sydney rock oysters (Saccostrea glomerata) Comp Biochem Physiol B. 2007;148:470–480. - PubMed
-
- Broehan G, Kemper M, Driemeier D, Vogelpohl I, Merzendorfer H. Cloning and expression analysis of midgut chymotrypsin-like proteinases in the tobacco hornworm. J Insect Physiol. 2008;54:1243–1252. - PubMed
-
- Broehan G, Arakane Y, Beeman RW, Kramer KJ, Muthukrishnan S, Merzendorfer H. Chymotrypsin-like peptidases from Tribolium castaneum: A role in molting revealed by RNA interference. Insect Biochem Mol Biol. 2010;40:274–283. - PubMed
-
- Cerenius L, Söderhäll K. The prophenoloxidase-activating system in invertebrates. Immunol Rev. 2004;1981:16–26. - PubMed
Internet Resources
-
- ORF Finder, http://www.ncbi.nlm.nih.gov/gorf (July 3, 2013).
-
- NCBI, http://www.ncbi.nlm.nih.gov (July 3, 2013).
-
- Expasy, http://www.expasy.ch/ (July 3, 2013).
-
- SignalP 4.0 software, http://www.cbs.dtu.dk/services/SignalP (July 3, 2013).
-
- ClustalW, http://www.ebi.ac.uk/Tools/msa/clustalw2/.
LinkOut - more resources
Full Text Sources
Other Literature Sources
