Mechanistic insights into the allosteric modulation of opioid receptors by sodium ions
- PMID: 25073009
- PMCID: PMC4131901
- DOI: 10.1021/bi5006915
Mechanistic insights into the allosteric modulation of opioid receptors by sodium ions
Abstract
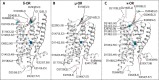
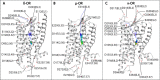
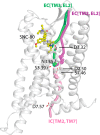
The idea of sodium ions altering G-protein-coupled receptor (GPCR) ligand binding and signaling was first suggested for opioid receptors (ORs) in the 1970s and subsequently extended to other GPCRs. Recently published ultra-high-resolution crystal structures of GPCRs, including that of the δ-OR subtype, have started to shed light on the mechanism underlying sodium control in GPCR signaling by revealing details of the sodium binding site. Whether sodium accesses different receptor subtypes from the extra- or intracellular sides, following similar or different pathways, is still an open question. Earlier experiments in brain homogenates suggested a differential sodium regulation of ligand binding to the three major OR subtypes, in spite of their high degree of sequence similarity. Intrigued by this possibility, we explored the dynamic nature of sodium binding to δ-OR, μ-OR, and κ-OR by means of microsecond-scale, all-atom molecular dynamics (MD) simulations. Rapid sodium permeation was observed exclusively from the extracellular milieu, and following similar binding pathways in all three ligand-free OR systems, notwithstanding extra densities of sodium observed near nonconserved residues of κ-OR and δ-OR, but not in μ-OR. We speculate that these differences may be responsible for the differential increase in antagonist binding affinity of μ-OR by sodium resulting from specific ligand binding experiments in transfected cells. On the other hand, sodium reduced the level of binding of subtype-specific agonists to all OR subtypes. Additional biased and unbiased MD simulations were conducted using the δ-OR ultra-high-resolution crystal structure as a model system to provide a mechanistic explanation for this experimental observation.
Figures

Similar articles
-
Pharmacologic Evidence for a Putative Conserved Allosteric Site on Opioid Receptors.Mol Pharmacol. 2018 Feb;93(2):157-167. doi: 10.1124/mol.117.109561. Epub 2017 Dec 12. Mol Pharmacol. 2018. PMID: 29233847 Free PMC article.
-
Exploring the structure of opioid receptors with homology modeling based on single and multiple templates and subsequent docking: a comparative study.J Mol Model. 2011 May;17(5):1207-21. doi: 10.1007/s00894-010-0803-8. Epub 2010 Jul 27. J Mol Model. 2011. PMID: 20661609
-
Comparative modeling and molecular dynamics studies of the delta, kappa and mu opioid receptors.Protein Eng. 1997 Sep;10(9):1019-38. doi: 10.1093/protein/10.9.1019. Protein Eng. 1997. PMID: 9464566
-
Opioid receptor types and subtypes: the delta receptor as a model.Annu Rev Pharmacol Toxicol. 1996;36:379-401. doi: 10.1146/annurev.pa.36.040196.002115. Annu Rev Pharmacol Toxicol. 1996. PMID: 8725395 Review.
-
[Structure of mu and delta opioid receptors].Med Sci (Paris). 2012 Oct;28(10):870-5. doi: 10.1051/medsci/20122810016. Epub 2012 Oct 12. Med Sci (Paris). 2012. PMID: 23067419 Review. French.
Cited by
-
An overview of recent molecular dynamics applications as medicinal chemistry tools for the undruggable site challenge.Medchemcomm. 2018 Apr 19;9(6):920-936. doi: 10.1039/c8md00166a. eCollection 2018 Jun 1. Medchemcomm. 2018. PMID: 30108981 Free PMC article. Review.
-
G-protein coupled receptors: advances in simulation and drug discovery.Curr Opin Struct Biol. 2016 Dec;41:83-89. doi: 10.1016/j.sbi.2016.06.008. Epub 2016 Jun 22. Curr Opin Struct Biol. 2016. PMID: 27344006 Free PMC article. Review.
-
Allostery at opioid receptors: modulation with small molecule ligands.Br J Pharmacol. 2018 Jul;175(14):2846-2856. doi: 10.1111/bph.13823. Epub 2017 Jun 7. Br J Pharmacol. 2018. PMID: 28419415 Free PMC article. Review.
-
Computational approaches to detect allosteric pathways in transmembrane molecular machines.Biochim Biophys Acta. 2016 Jul;1858(7 Pt B):1652-62. doi: 10.1016/j.bbamem.2016.01.010. Epub 2016 Jan 22. Biochim Biophys Acta. 2016. PMID: 26806157 Free PMC article.
-
G protein signaling-biased mu opioid receptor agonists that produce sustained G protein activation are noncompetitive agonists.Proc Natl Acad Sci U S A. 2021 Nov 30;118(48):e2102178118. doi: 10.1073/pnas.2102178118. Proc Natl Acad Sci U S A. 2021. PMID: 34819362 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

