Control of RecBCD enzyme activity by DNA binding- and Chi hotspot-dependent conformational changes
- PMID: 25073102
- PMCID: PMC4188757
- DOI: 10.1016/j.jmb.2014.07.017
Control of RecBCD enzyme activity by DNA binding- and Chi hotspot-dependent conformational changes
Abstract
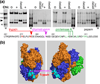
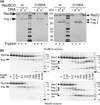
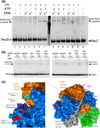
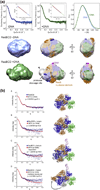
Faithful repair of DNA double-strand breaks by homologous recombination is crucial to maintain functional genomes. The major Escherichia coli pathway of DNA break repair requires RecBCD enzyme, a complex protein machine with multiple activities. Upon encountering a Chi recombination hotspot (5' GCTGGTGG 3') during DNA unwinding, RecBCD's unwinding, nuclease, and RecA-loading activities change dramatically, but the physical basis for these changes is unknown. Here, we identify, during RecBCD's DNA unwinding, two Chi-stimulated conformational changes involving RecC. One produced a marked, long-lasting, Chi-dependent increase in protease sensitivity of a small patch, near the Chi recognition domain, on the solvent-exposed RecC surface. The other change was identified by crosslinking of an artificial amino acid inserted in this RecC patch to RecB. Small-angle X-ray scattering analysis confirmed a major conformational change upon binding of DNA to the enzyme and is consistent with these two changes. We propose that, upon DNA binding, the RecB nuclease domain swings from one side of RecC to the other; when RecBCD encounters Chi, the nuclease domain returns to its initial position determined by crystallography, where it nicks DNA exiting from RecC and loads RecA onto the newly generated 3'-ended single-stranded DNA during continued unwinding; a crevice between RecB and RecC increasingly narrows during these steps. This model provides a physical basis for the intramolecular "signal transduction" from Chi to RecC to RecD to RecB inferred previously from genetic and enzymatic analyses, and it accounts for the enzymatic changes that accompany Chi's stimulation of recombination.
Keywords: SAXS; crosslinking; helicase–nuclease; limited proteolysis; recombination.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Figures






Similar articles
-
RecBCD enzyme: mechanistic insights from mutants of a complex helicase-nuclease.Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0004123. doi: 10.1128/mmbr.00041-23. Epub 2023 Dec 4. Microbiol Mol Biol Rev. 2023. PMID: 38047637 Free PMC article. Review.
-
RecBCD Enzyme "Chi Recognition" Mutants Recognize Chi Recombination Hotspots in the Right DNA Context.Genetics. 2016 Sep;204(1):139-52. doi: 10.1534/genetics.116.191056. Epub 2016 Jul 8. Genetics. 2016. PMID: 27401752 Free PMC article.
-
Chi hotspot Control of RecBCD Helicase-nuclease: Enzymatic Tests Support the Intramolecular Signal-transduction Model.J Mol Biol. 2024 Mar 15;436(6):168482. doi: 10.1016/j.jmb.2024.168482. Epub 2024 Feb 7. J Mol Biol. 2024. PMID: 38331210 Free PMC article.
-
A flexible RecC surface loop required for Chi hotspot control of RecBCD enzyme.Genetics. 2023 Mar 2;223(3):iyac175. doi: 10.1093/genetics/iyac175. Genetics. 2023. PMID: 36521180 Free PMC article.
-
Chi and the RecBC D enzyme of Escherichia coli.Annu Rev Genet. 1994;28:49-70. doi: 10.1146/annurev.ge.28.120194.000405. Annu Rev Genet. 1994. PMID: 7893137 Review.
Cited by
-
The positioning of Chi sites allows the RecBCD pathway to suppress some genomic rearrangements.Nucleic Acids Res. 2019 Feb 28;47(4):1836-1846. doi: 10.1093/nar/gky1252. Nucleic Acids Res. 2019. PMID: 30544167 Free PMC article.
-
How Does a Helicase Unwind DNA? Insights from RecBCD Helicase.Bioessays. 2018 Jun;40(6):e1800009. doi: 10.1002/bies.201800009. Epub 2018 Mar 30. Bioessays. 2018. PMID: 29603305 Free PMC article. Review.
-
RecBCD enzyme: mechanistic insights from mutants of a complex helicase-nuclease.Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0004123. doi: 10.1128/mmbr.00041-23. Epub 2023 Dec 4. Microbiol Mol Biol Rev. 2023. PMID: 38047637 Free PMC article. Review.
-
An Hfq-dependent post-transcriptional mechanism fine tunes RecB expression in Escherichia coli.Elife. 2025 Aug 12;13:RP94918. doi: 10.7554/eLife.94918. Elife. 2025. PMID: 40794102 Free PMC article.
-
ATP hydrolysis provides functions that promote rejection of pairings between different copies of long repeated sequences.Nucleic Acids Res. 2017 Aug 21;45(14):8448-8462. doi: 10.1093/nar/gkx582. Nucleic Acids Res. 2017. PMID: 28854739 Free PMC article.
References
-
- Ganesan S, Smith GR. Strand-specific binding to duplex DNA ends by the subunits of Escherichia coli RecBCD enzyme. J Mol Biol. 1993;229:67–78. - PubMed
-
- Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. - PubMed
-
- Taylor A, Smith GR. Unwinding and rewinding of DNA by the RecBC enzyme. Cell. 1980;22:447–457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

