Genome-wide analysis of histone modifications in human endometrial stromal cells
- PMID: 25073104
- PMCID: PMC5414786
- DOI: 10.1210/me.2014-1117
Genome-wide analysis of histone modifications in human endometrial stromal cells
Abstract
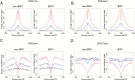
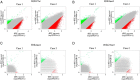
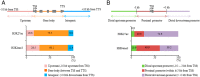
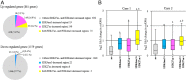
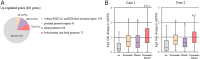
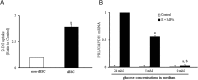
Dramatic changes of gene expressions occur in human endometrial stromal cells (ESCs) during decidualization. The changes in gene expression are associated with changes of chromatin structure, which are regulated by histone modifications. Here we investigated genome-wide changes in histone modifications associated with decidualization in human ESCs using chromatin immunoprecipitation combined with next-generation sequencing. ESCs were incubated with estradiol and medroxyprogesterone acetate for 14 days to induce decidualization. The chromatin immunoprecipitation-sequence data showed that induction of decidualization increased H3K27ac and H3K4me3 signals in many genomic regions but decreased in only a few regions. Most of the H3K27ac-increased regions (80%) and half of the H3K4me3-increased regions were located in the distal promoter regions (more than 3 kb upstream or downstream of the transcription start site). RNA sequence showed that induction of decidualization up-regulated 881 genes, 223 of which had H3K27ac- or H3K4me3-increased regions in the proximal and distal promoter regions. Induction of decidualization increased the mRNA levels of these genes more than it increased the mRNA levels of genes without H3K27ac- or H3K4me3-increased regions. Pathway analysis revealed that up-regulated genes with the H3K27ac- or H3K4me3-increased regions were associated with the insulin signaling, which may be involved in glucose uptake that is necessary for ESCs to undergo decidualization. These results show that histone modification statuses on a genome-wide basis change in human ESCs during decidualization. The main changes of histone modifications are increases of H3K27ac and H3K4me3 in both the proximal and distal promoter regions, which are involved in the up-regulation of gene expression that occurs during decidualization.
Figures

Similar articles
-
Transcription factor C/EBPβ induces genome-wide H3K27ac and upregulates gene expression during decidualization of human endometrial stromal cells.Mol Cell Endocrinol. 2021 Jan 15;520:111085. doi: 10.1016/j.mce.2020.111085. Epub 2020 Nov 21. Mol Cell Endocrinol. 2021. PMID: 33232782
-
Genome-wide DNA methylation analysis revealed stable DNA methylation status during decidualization in human endometrial stromal cells.BMC Genomics. 2019 Apr 29;20(1):324. doi: 10.1186/s12864-019-5695-0. BMC Genomics. 2019. PMID: 31035926 Free PMC article.
-
Importance of C/EBPβ binding and histone acetylation status in the promoter regions for induction of IGFBP-1, PRL, and Mn-SOD by cAMP in human endometrial stromal cells.Endocrinology. 2014 Jan;155(1):275-86. doi: 10.1210/en.2013-1569. Epub 2013 Dec 20. Endocrinology. 2014. PMID: 24248464
-
Genome-wide analysis of histone modifications that underlie the dynamic changes in gene expression during decidualization in human endometrial stromal cells.Mol Hum Reprod. 2023 Jun 30;29(7):gaad019. doi: 10.1093/molehr/gaad019. Mol Hum Reprod. 2023. PMID: 37310913 Review.
-
Glucose regulates the histone acetylation of gene promoters in decidualizing stromal cells.Reproduction. 2019 May;157(5):457-464. doi: 10.1530/REP-18-0393. Reproduction. 2019. PMID: 30817321
Cited by
-
The Regulators of Human Endometrial Stromal Cell Decidualization.Biomolecules. 2022 Sep 10;12(9):1275. doi: 10.3390/biom12091275. Biomolecules. 2022. PMID: 36139114 Free PMC article. Review.
-
H3K27me3 dynamics dictate evolving uterine states in pregnancy and parturition.J Clin Invest. 2018 Jan 2;128(1):233-247. doi: 10.1172/JCI95937. Epub 2017 Nov 27. J Clin Invest. 2018. PMID: 29202469 Free PMC article.
-
Histone acetylation and the role of histone deacetylases in normal cyclic endometrium.Reprod Biol Endocrinol. 2020 Aug 13;18(1):84. doi: 10.1186/s12958-020-00637-5. Reprod Biol Endocrinol. 2020. PMID: 32791974 Free PMC article. Review.
-
Concurrent mapping of multiple epigenetic marks and co-occupancy using ACT2-seq.Cell Biosci. 2021 Dec 4;11(1):198. doi: 10.1186/s13578-021-00711-4. Cell Biosci. 2021. PMID: 34863284 Free PMC article.
-
Differential gene expression in decidualized human endometrial stromal cells induced by different stimuli.Sci Rep. 2024 Apr 2;14(1):7726. doi: 10.1038/s41598-024-58065-z. Sci Rep. 2024. PMID: 38565619 Free PMC article.
References
-
- Gellersen B, Brosens J. Cyclic AMP and progesterone receptor cross-talk in human endometrium: a decidualizing affair. J Endocrinol. 2003;178:357–372. - PubMed
-
- Laird SM, Tuckerman EM, Li TC. Cytokine expression in the endometrium of women with implantation failure and recurrent miscarriage. Reprod Biomed Online. 2006;13:13–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

