Antivirulence activity of the human gut metabolome
- PMID: 25073640
- PMCID: PMC4128352
- DOI: 10.1128/mBio.01183-14
Antivirulence activity of the human gut metabolome
Abstract
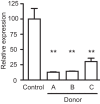
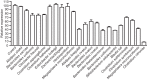
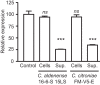
The mammalian gut contains a complex assembly of commensal microbes termed microbiota. Although much has been learned about the role of these microbes in health, the mechanisms underlying these functions are ill defined. We have recently shown that the mammalian gut contains thousands of small molecules, most of which are currently unidentified. Therefore, we hypothesized that these molecules function as chemical cues used by hosts and microbes during their interactions in health and disease. Thus, a search was initiated to identify molecules produced by the microbiota that are sensed by pathogens. We found that a secreted molecule produced by clostridia acts as a strong repressor of Salmonella virulence, obliterating expression of the Salmonella pathogenicity island 1 as well as host cell invasion. It has been known for decades that the microbiota protects its hosts from invading pathogens, and these data suggest that chemical sensing may be involved in this phenomenon. Further investigations should reveal the exact biological role of this molecule as well as its therapeutic potential. Importance: Microbes can communicate through the production and sensing of small molecules. Within the complex ecosystem formed by commensal microbes living in and on the human body, it is likely that these molecular messages are used extensively during the interactions between different microbial species as well as with host cells. Deciphering such a molecular dialect will be fundamental to our understanding of host-microbe interactions in health and disease and may prove useful for the design of new therapeutic strategies that target these mechanisms of communication.
Copyright © 2014 Antunes et al.
Figures

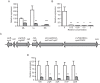
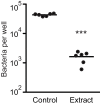
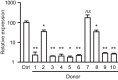
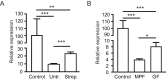

References
-
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U. S. A. 110:3229–3236. 10.1073/pnas.1218525110 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

