Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution
- PMID: 25073643
- PMCID: PMC4128362
- DOI: 10.1128/mBio.01494-14
Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution
Abstract
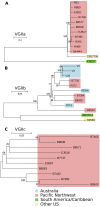
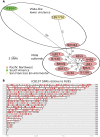
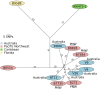
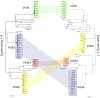
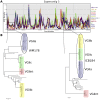
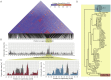
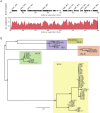
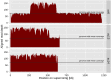
An outbreak of the fungal pathogen Cryptococcus gattii began in the Pacific Northwest (PNW) in the late 1990s. This outbreak consists of three clonal subpopulations: VGIIa/major, VGIIb/minor, and VGIIc/novel. Both VGIIa and VGIIc are unique to the PNW and exhibit increased virulence. In this study, we sequenced the genomes of isolates from these three groups, as well as global isolates, and analyzed a total of 53 isolates. We found that VGIIa/b/c populations show evidence of clonal expansion in the PNW. Whole-genome sequencing provided evidence that VGIIb originated in Australia, while VGIIa may have originated in South America, and these were likely independently introduced. Additionally, the VGIIa outbreak lineage may have arisen from a less virulent clade that contained a mutation in the MSH2 ortholog, but this appears to have reverted in the VGIIa outbreak strains, suggesting that a transient mutator phenotype may have contributed to adaptation and evolution of virulence in the PNW outbreak. PNW outbreak isolates share genomic islands, both between the clonal lineages and with global isolates, indicative of sexual recombination. This suggests that VGII C. gattii has undergone sexual reproduction, either bisexual or unisexual, in multiple locales contributing to the production of novel, virulent subtypes. We also found that the genomes of two basal VGII isolates from HIV(+) patients contain an introgression tract spanning three genes. Introgression substantially contributed to intra-VGII polymorphism and likely occurred through sexual reproduction with VGI. More broadly, these findings illustrate how both microevolution and sexual reproduction play central roles in the development of infectious outbreaks from avirulent or less virulent progenitors. Importance: Cryptococcus gattii is the causative agent responsible for ongoing infections in the Pacific Northwest of the United States and western Canada. The incidence of these infections increased dramatically in the 1990s and remains elevated. These infections are attributable to three clonal lineages of C. gattii, VGIIa, VGIIb, and VGIIc, with only VGIIa identified once previously in the Pacific Northwest prior to the start of the outbreak, albeit in a less virulent form. This study addresses the origin and emergence of this outbreak, using whole-genome sequencing and comparison of both outbreak and global isolates. We show that VGIIa arose mitotically from a less virulent clonal group, possibly via the action of a mutator phenotype, while VGIIb was likely introduced from Australia, and VGIIc appears to have emerged in the United States or in an undersampled locale via sexual reproduction. This work shows that multiple processes can contribute to the emergence of an outbreak.
Copyright © 2014 Billmyre et al.
Figures


References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
