Suberoylanilide hydroxamic acid (SAHA)-induced dynamics of a human histone deacetylase protein interaction network
- PMID: 25073741
- PMCID: PMC4223495
- DOI: 10.1074/mcp.M113.037127
Suberoylanilide hydroxamic acid (SAHA)-induced dynamics of a human histone deacetylase protein interaction network
Abstract
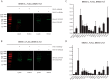
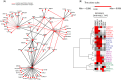
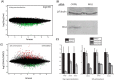
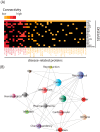
Histone deacetylases (HDACs) are targets for cancer therapy. Suberoylanilide hydroxamic acid (SAHA) is an HDAC inhibitor approved by the U.S. Food and Drug Administration for the treatment of cutaneous T-cell lymphoma. To obtain a better mechanistic understanding of the Sin3/HDAC complex in cancer, we extended its protein-protein interaction network and identified a mutually exclusive pair within the complex. We then assessed the effects of SAHA on the disruption of the complex network through six homologous baits. SAHA perturbs multiple protein interactions and therefore compromises the composition of large parts of the Sin3/HDAC network. A comparison of the effect of SAHA treatment on gene expression in breast cancer cells to a knockdown of the ING2 subunit indicated that a portion of the anticancer effects of SAHA may be attributed to the disruption of ING2's association with the complex. Our dynamic protein interaction network resource provides novel insights into the molecular mechanism of SAHA action and demonstrates the potential for drugs to rewire networks.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

