Strand-specific (asymmetric) contribution of phosphodiester linkages on RNA polymerase II transcriptional efficiency and fidelity
- PMID: 25074911
- PMCID: PMC4136571
- DOI: 10.1073/pnas.1406234111
Strand-specific (asymmetric) contribution of phosphodiester linkages on RNA polymerase II transcriptional efficiency and fidelity
Abstract
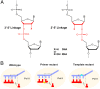
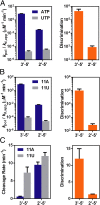
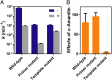
Nonenzymatic RNA polymerization in early life is likely to introduce backbone heterogeneity with a mixture of 2'-5' and 3'-5' linkages. On the other hand, modern nucleic acids are dominantly composed of 3'-5' linkages. RNA polymerase II (pol II) is a key modern enzyme responsible for synthesizing 3'-5'-linked RNA with high fidelity. It is not clear how modern enzymes, such as pol II, selectively recognize 3'-5' linkages over 2'-5' linkages of nucleic acids. In this work, we systematically investigated how phosphodiester linkages of nucleic acids govern pol II transcriptional efficiency and fidelity. Through dissecting the impacts of 2'-5' linkage mutants in the pol II catalytic site, we revealed that the presence of 2'-5' linkage in RNA primer only modestly reduces pol II transcriptional efficiency without affecting pol II transcriptional fidelity. In sharp contrast, the presence of 2'-5' linkage in DNA template leads to dramatic decreases in both transcriptional efficiency and fidelity. These distinct effects reveal that pol II has an asymmetric (strand-specific) recognition of phosphodiester linkage. Our results provided important insights into pol II transcriptional fidelity, suggesting essential contributions of phosphodiester linkage to pol II transcription. Finally, our results also provided important understanding on the molecular basis of nucleic acid recognition and genetic information transfer during molecular evolution. We suggest that the asymmetric recognition of phosphodiester linkage by modern nucleic acid enzymes likely stems from the distinct evolutionary pressures of template and primer strand in genetic information transfer during molecular evolution.
Keywords: 2′–5′ phosphodiester linkage; synthetic nucleic acid analogues; trigger loop.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


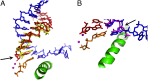
Similar articles
-
Impact of template backbone heterogeneity on RNA polymerase II transcription.Nucleic Acids Res. 2015 Feb 27;43(4):2232-41. doi: 10.1093/nar/gkv059. Epub 2015 Feb 6. Nucleic Acids Res. 2015. PMID: 25662224 Free PMC article.
-
Dissecting chemical interactions governing RNA polymerase II transcriptional fidelity.J Am Chem Soc. 2012 May 16;134(19):8231-40. doi: 10.1021/ja302077d. Epub 2012 May 2. J Am Chem Soc. 2012. PMID: 22509745 Free PMC article.
-
Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis.Cell. 2006 Dec 1;127(5):941-54. doi: 10.1016/j.cell.2006.11.023. Cell. 2006. PMID: 17129781 Free PMC article.
-
RNA polymerase II transcriptional fidelity control and its functional interplay with DNA modifications.Crit Rev Biochem Mol Biol. 2015;50(6):503-19. doi: 10.3109/10409238.2015.1087960. Epub 2015 Sep 22. Crit Rev Biochem Mol Biol. 2015. PMID: 26392149 Free PMC article. Review.
-
Molecular basis of transcriptional fidelity and DNA lesion-induced transcriptional mutagenesis.DNA Repair (Amst). 2014 Jul;19:71-83. doi: 10.1016/j.dnarep.2014.03.024. Epub 2014 Apr 21. DNA Repair (Amst). 2014. PMID: 24767259 Free PMC article. Review.
Cited by
-
Impact of template backbone heterogeneity on RNA polymerase II transcription.Nucleic Acids Res. 2015 Feb 27;43(4):2232-41. doi: 10.1093/nar/gkv059. Epub 2015 Feb 6. Nucleic Acids Res. 2015. PMID: 25662224 Free PMC article.
-
Functional interplay between NTP leaving group and base pair recognition during RNA polymerase II nucleotide incorporation revealed by methylene substitution.Nucleic Acids Res. 2016 May 5;44(8):3820-8. doi: 10.1093/nar/gkw220. Epub 2016 Apr 7. Nucleic Acids Res. 2016. PMID: 27060150 Free PMC article.
-
Elucidation of the Dynamics of Transcription Elongation by RNA Polymerase II using Kinetic Network Models.Acc Chem Res. 2016 Apr 19;49(4):687-94. doi: 10.1021/acs.accounts.5b00536. Epub 2016 Mar 18. Acc Chem Res. 2016. PMID: 26991064 Free PMC article.
-
Functional assays for transcription mechanisms in high-throughput.Methods. 2019 Apr 15;159-160:115-123. doi: 10.1016/j.ymeth.2019.02.017. Epub 2019 Feb 20. Methods. 2019. PMID: 30797033 Free PMC article. Review.
-
The m6A methylation perturbs the Hoogsteen pairing-guided incorporation of an oxidized nucleotide.Chem Sci. 2017 Sep 1;8(9):6380-6388. doi: 10.1039/c7sc02340e. Epub 2017 Jul 6. Chem Sci. 2017. PMID: 29308175 Free PMC article.
References
-
- Gilbert W. Origin of life: The RNA world. Nature. 1986;319(6055):618.
-
- Joyce GF. RNA evolution and the origins of life. Nature. 1989;338(6212):217–224. - PubMed
-
- Ekland EH, Bartel DP. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature. 1996;382(6589):373–376. - PubMed
-
- Ertem G, Ferris JP. Synthesis of RNA oligomers on heterogeneous templates. Nature. 1996;379(6562):238–240. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

