Enrichment Map - a Cytoscape app to visualize and explore OMICs pathway enrichment results
- PMID: 25075306
- PMCID: PMC4103489
- DOI: 10.12688/f1000research.4536.1
Enrichment Map - a Cytoscape app to visualize and explore OMICs pathway enrichment results
Abstract
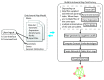
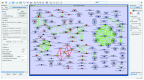
High-throughput OMICs experiments generate signals for millions of entities (i.e. genes, proteins, metabolites or any measurable biological entity) in the cell. In an effort to summarize and explore these signals, expression results are examined in the context of known pathways and processes, through enrichment analysis to generate a set of pathways and processes that is significantly enriched. Due to the high redundancy in annotation resources this often results in hundreds of sets. To facilitate the analysis of these results, we have developed the Enrichment Map app to visualize enrichments as a network. We have updated Enrichment Map to support Cytoscape 3, and have added additional features including new data formats and command line access.
Conflict of interest statement
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

