Improving prediction of type 1 diabetes by testing non-HLA genetic variants in addition to HLA markers
- PMID: 25075402
- PMCID: PMC4116638
- DOI: 10.1111/pedi.12092
Improving prediction of type 1 diabetes by testing non-HLA genetic variants in addition to HLA markers
Abstract
Objective: The purpose of this study was to explore whether non-human leukocyte antigen (non-HLA) genetic markers can improve type 1 diabetes(T1D) prediction in a prospective cohort with high-risk HLA-DR,DQ genotypes.
Methods: The Diabetes Autoimmunity Study in the Young (DAISY) follows prospectively for the development of T1D and islet autoimmunity (IA)children at increased genetic risk. A total of 1709 non-Hispanic White DAISY participants have been genotyped for 27 non-HLA single nucleotide polymorphisms (SNPs) and one microsatellite.
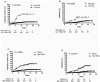
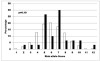
Results: In multivariate analyses adjusting for family history and HLA-DR3/4 genotype, PTPN22 (rs2476601) and two UBASH3A (rs11203203 and rs9976767) SNPs were associated with development of IA [hazard ratio(HR)=1.87, 1.55, and 1.54, respectively, all p ≤ 0.003], while GLIS3 and IL2RA showed borderline association with development of IA. INS,UBASH3A, and IFIH1 were significantly associated with progression from IA to diabetes (HR=1.65, 1.44, and 1.47, respectively, all p ≤ 0.04), while PTPN22 and IL27 showed borderline association with progression from IA to diabetes. In survival analysis, 45% of general population DAISY children with PTPN22 rs2476601 TT or HLA-DR3/4 and UBASH3A rs11203203 AA developed diabetes by age 15, compared with 3% of children with all other genotypes (p<0.0001). Addition of non-HLA markers to HLA-DR3/4,DQ8 did not improve diabetes prediction in first-degree relatives.
Conclusion: Addition of PTPN22 and UBASH3A SNPs to HLA-DR,DQ genotyping can improve T1D risk prediction.
Keywords: islet autoimmunity; non-HLA genetic markers; prediction; type 1 diabetes.
2013 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures

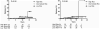
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

