Deep mutational scanning: a new style of protein science
- PMID: 25075907
- PMCID: PMC4410700
- DOI: 10.1038/nmeth.3027
Deep mutational scanning: a new style of protein science
Abstract
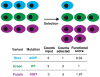
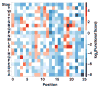
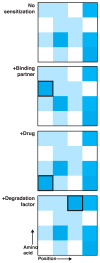
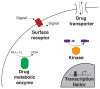
Mutagenesis provides insight into proteins, but only recently have assays that couple genotype to phenotype been used to assess the activities of as many as 1 million mutant versions of a protein in a single experiment. This approach-'deep mutational scanning'-yields large-scale data sets that can reveal intrinsic protein properties, protein behavior within cells and the consequences of human genetic variation. Deep mutational scanning is transforming the study of proteins, but many challenges must be tackled for it to fulfill its promise.
Conflict of interest statement
The authors have no competing financial interests to declare.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

