Epigenetic dysregulation: a novel pathway of oncogenesis in pediatric brain tumors
- PMID: 25077668
- PMCID: PMC4201756
- DOI: 10.1007/s00401-014-1325-8
Epigenetic dysregulation: a novel pathway of oncogenesis in pediatric brain tumors
Abstract
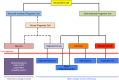
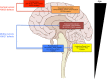
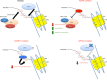
A remarkably large number of "epigenetic regulators" have been recently identified to be altered in cancers and a rapidly expanding body of literature points to "epigenetic addiction" (an aberrant epigenetic state to which a tumor is addicted) as a new previously unsuspected mechanism of oncogenesis. Although mutations are also found in canonical signaling pathway genes, we and others identified chromatin-associated proteins to be more commonly altered by somatic alterations than any other class of oncoprotein in several subgroups of childhood high-grade brain tumors. Furthermore, as these childhood malignancies carry fewer non-synonymous somatic mutations per case in contrast to most adult cancers, these mutations are likely drivers in these tumors. Herein, we will use as examples of this novel hallmark of oncogenesis high-grade astrocytomas, including glioblastoma, and a subgroup of embryonal tumors, embryonal tumor with multilayered rosettes (ETMR) to describe the novel molecular defects uncovered in these deadly tumors. We will further discuss evidence for their profound effects on the epigenome. The relative genetic simplicity of these tumors promises general insights into how mutations in the chromatin machinery modify downstream epigenetic signatures to drive transformation, and how to target this plastic genetic/epigenetic interface.
Figures

References
-
- Al-Halabi H, Nantel A, Klekner A, Guiot MC, Albrecht S, Hauser P, Garami M, Bognar L, Kavan P, Gerges N, Shirinian M, Roberge D, Muanza T, Jabado N. Preponderance of sonic hedgehog pathway activation characterizes adult medulloblastoma. Acta Neuropathol. 2011;121(2):229–239. doi: 10.1007/s00401-010-0780-0. - DOI - PubMed
-
- Becher OJ, Hambardzumyan D, Walker TR, Helmy K, Nazarian J, Albrecht S, Hiner RL, Gall S, Huse JT, Jabado N, MacDonald TJ, Holland EC. Preclinical evaluation of radiation and perifosine in a genetically and histologically accurate model of brainstem glioma. Cancer Res. 2010;70(6):2548–2557. doi: 10.1158/0008-5472.CAN-09-2503. - DOI - PMC - PubMed
-
- Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H, Nik-Zainal S, Martin S, McLaren S, Goody V, Robinson B, Butler A, Teague JW, Halai D, Khatri B, Myklebost O, Baumhoer D, Jundt G, Hamoudi R, Tirabosco R, Amary MF, Futreal PA, Stratton MR, Campbell PJ, Flanagan AM. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45(12):1479–1482. doi: 10.1038/ng.2814. - DOI - PMC - PubMed
-
- Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, Radlwimmer B, Hojfeldt JW, Truffaux N, Castel D, Schubert S, Ryzhova M, Seker-Cin H, Gronych J, Johann PD, Stark S, Meyer J, Milde T, Schuhmann M, Ebinger M, Monoranu CM, Ponnuswami A, Chen S, Jones C, Witt O, Collins VP, von Deimling A, Jabado N, Puget S, Grill J, Helin K, Korshunov A, Lichter P, Monje M, Plass C, Cho YJ, Pfister SM. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24(5):660–672. doi: 10.1016/j.ccr.2013.10.006. - DOI - PubMed
-
- Bjerke L, Mackay A, Nandhabalan M, Burford A, Jury A, Popov S, Bax DA, Carvalho D, Taylor KR, Vinci M, Bajrami I, McGonnell IM, Lord CJ, Reis RM, Hargrave D, Ashworth A, Workman P, Jones C (2013) Histone H3.3 mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov. doi:10.1158/2159-8290.CD-12-0426 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

