MicroRNA regulation of mitogenic signaling networks in the human placenta
- PMID: 25077964
- PMCID: PMC4215224
- DOI: 10.1074/jbc.M114.587295
MicroRNA regulation of mitogenic signaling networks in the human placenta
Abstract
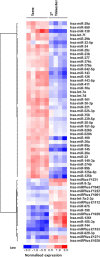
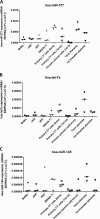
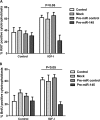
Placental cell growth depends on an adaptable combination of an endogenous developmental program and the exogenous influence of maternal growth factors, both of which may be influenced by microRNA (miR)-dependent effects on gene expression. We have previously shown that global miR suppression in placenta accelerates proliferation and enhances levels of growth factor signaling mediators in cytotrophoblast. This study aimed to identify miRs involved in regulating placental growth. An initial array revealed 58 miR species whose expression differs between first trimester, when cytotrophoblast proliferation is rapid, and term, by which time proliferation has slowed. In silico analysis defined potential growth-regulatory miRs; among these, hsa-miR-145, hsa-miR-377, and hsa-let-7a were predicted to target known placental growth genes and were higher at term than in the first trimester, so they were selected for further analysis. Overexpression of miR-377 and let-7a, but not miR-145, in first trimester placental explants significantly reduced basal cytotrophoblast proliferation and expression of ERK and MYC. PCR arrays, in silico analysis, Western blotting, and 3'-UTR luciferase reporter assays revealed targets of miR-145 within the insulin-like growth factor axis. Analysis of proliferation in placental explants overexpressing miR-145 demonstrated its role as a mediator of insulin-like growth factor-induced trophoblast proliferation. These findings identify miR-377 and let-7a in regulation of endogenous cell growth and miR-145 in the placental response to maternal stimulation and will aid the development of therapeutic strategies for problem pregnancies.
Keywords: Developmental Factor; Gene Regulation; Insulin-like Growth Factor (IGF); MicroRNA (miRNA); Placenta; Proliferation; Reproduction; Trophoblast.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Arnholdt H., Meisel F., Fandrey K., Löhrs U. (1991) Proliferation of villous trophoblast of the human placenta in normal and abnormal pregnancies. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 60, 365–372 - PubMed
-
- Jansson T., Powell T. L. (2006) Human placental transport in altered fetal growth: does the placenta function as a nutrient sensor? a review. Placenta 27, 91–97 - PubMed
-
- Forbes K., Westwood M., Baker P. N., Aplin J. D. (2008) Insulin-like growth factor I and II regulate the life cycle of trophoblast in the developing human placenta. Am. J. Physiol. Cell Physiol. 294, C1313–C1322 - PubMed
-
- Forbes K., Souquet B., Garside R., Aplin J. D., Westwood M. (2010) Transforming growth factor-β (TGFβ) receptors I/II differentially regulate TGFβ1 and IGF-binding protein-3 mitogenic effects in the human placenta. Endocrinology 151, 1723–1731 - PubMed
-
- Forbes K., West G., Garside R., Aplin J. D., Westwood M. (2009) The protein-tyrosine phosphatase, SRC homology-2 domain containing protein tyrosine phosphatase-2, is a crucial mediator of exogenous insulin-like growth factor signaling to human trophoblast. Endocrinology 150, 4744–4754 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

