Analysis of culture-dependent versus culture-independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid
- PMID: 25078910
- PMCID: PMC4187760
- DOI: 10.1128/JCM.01028-14
Analysis of culture-dependent versus culture-independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid
Abstract
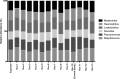
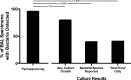
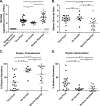
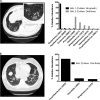
The diagnosis and management of pneumonia are limited by the use of culture-based techniques of microbial identification, which may fail to identify unculturable, fastidious, and metabolically active viable but unculturable bacteria. Novel high-throughput culture-independent techniques hold promise but have not been systematically compared to conventional culture. We analyzed 46 clinically obtained bronchoalveolar lavage (BAL) fluid specimens from symptomatic and asymptomatic lung transplant recipients both by culture (using a clinical microbiology laboratory protocol) and by bacterial 16S rRNA gene pyrosequencing. Bacteria were identified in 44 of 46 (95.7%) BAL fluid specimens by culture-independent sequencing, significantly more than the number of specimens in which bacteria were detected (37 of 46, 80.4%, P ≤ 0.05) or "pathogen" species reported (18 of 46, 39.1%, P ≤ 0.0001) via culture. Identification of bacteria by culture was positively associated with culture-independent indices of infection (total bacterial DNA burden and low bacterial community diversity) (P ≤ 0.01). In BAL fluid specimens with no culture growth, the amount of bacterial DNA was greater than that in reagent and rinse controls, and communities were markedly dominated by select Gammaproteobacteria, notably Escherichia species and Pseudomonas fluorescens. Culture growth above the threshold of 10(4) CFU/ml was correlated with increased bacterial DNA burden (P < 0.01), decreased community diversity (P < 0.05), and increased relative abundance of Pseudomonas aeruginosa (P < 0.001). We present two case studies in which culture-independent techniques identified a respiratory pathogen missed by culture and clarified whether a cultured "oral flora" species represented a state of acute infection. In summary, we found that bacterial culture of BAL fluid is largely effective in discriminating acute infection from its absence and identified some specific limitations of BAL fluid culture in the diagnosis of pneumonia. We report the first correlation of quantitative BAL fluid culture results with culture-independent evidence of infection.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Hoyert DL, Xu J. 2012. Deaths: preliminary data for 2011. Natl. Vital Stat. Rep. 61:1–65 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

