Bacteriophages as vehicles for antibiotic resistance genes in the environment
- PMID: 25078987
- PMCID: PMC4117541
- DOI: 10.1371/journal.ppat.1004219
Bacteriophages as vehicles for antibiotic resistance genes in the environment
Conflict of interest statement
The author has declared that no competing interests exist.
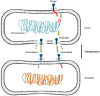
Figures

References
-
- Marti E, Variatza E, Balcazar JL (2014) The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol 22: 36–41. - PubMed
-
- Frost LS, Leplae R, Summers AO, Toussaint A (2005) Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3: 722–732. - PubMed
-
- Brabban AD, Hite E, Callaway TR (2005) Evolution of foodborne pathogens via temperate bacteriophage-mediated gene transfer. Foodborne Pathog Dis 2: 287–303. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

