Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity
- PMID: 25079331
- PMCID: PMC4184961
- DOI: 10.1038/nature13556
Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity
Abstract
Cancers arise through a process of somatic evolution that can result in substantial sub-clonal heterogeneity within tumours. The mechanisms responsible for the coexistence of distinct sub-clones and the biological consequences of this coexistence remain poorly understood. Here we used a mouse xenograft model to investigate the impact of sub-clonal heterogeneity on tumour phenotypes and the competitive expansion of individual clones. We found that tumour growth can be driven by a minor cell subpopulation, which enhances the proliferation of all cells within a tumour by overcoming environmental constraints and yet can be outcompeted by faster proliferating competitors, resulting in tumour collapse. We developed a mathematical modelling framework to identify the rules underlying the generation of intra-tumour clonal heterogeneity. We found that non-cell-autonomous driving of tumour growth, together with clonal interference, stabilizes sub-clonal heterogeneity, thereby enabling inter-clonal interactions that can lead to new phenotypic traits.
Figures

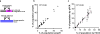

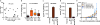
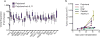
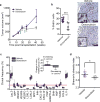


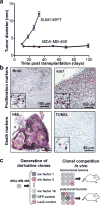



Comment in
-
Minor clone may drive cancer growth.Cancer Discov. 2014 Oct;4(10):1109-10. doi: 10.1158/2159-8290.CD-NB2014-126. Epub 2014 Aug 14. Cancer Discov. 2014. PMID: 25274666 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

