In Silico cancer cell versus stroma cellularity index computed from species-specific human and mouse transcriptome of xenograft models: towards accurate stroma targeting therapy assessment
- PMID: 25079962
- PMCID: PMC4101338
- DOI: 10.1186/1755-8794-7-S1-S2
In Silico cancer cell versus stroma cellularity index computed from species-specific human and mouse transcriptome of xenograft models: towards accurate stroma targeting therapy assessment
Abstract
Background: The current state of the art for measuring stromal response to targeted therapy requires burdensome and rate limiting quantitative histology. Transcriptome measures are increasingly affordable and provide an opportunity for developing a stromal versus cancer ratio in xenograft models. In these models, human cancer cells are transplanted into mouse host tissues (stroma) and together coevolve into a tumour microenvironment. However, profiling the mouse or human component separately remains problematic. Indeed, laser capture microdissection is labour intensive. Moreover, gene expression using commercial microarrays introduces significant and underreported cross-species hybridization errors that are commonly overlooked by biologists.
Method: We developed a customized dual-species array, H&M array, and performed cross-species and species-specific hybridization measurements. We validated a new methodology for establishing the stroma vs cancer ratio using transcriptomic data.
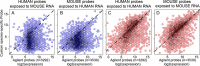
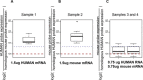
Results: In the biological validation of the H&M array, cross-species hybridization of human and mouse probes was significantly reduced (4.5 and 9.4 fold reduction, respectively; p < 2x10-16 for both, Mann-Whitney test). We confirmed the capability of the H&M array to determine the stromal to cancer cells ratio based on the estimation of cellularity index of mouse/human mRNA content in vitro. This new metrics enable to investigate more efficiently the stroma-cancer cell interactions (e.g. cellularity) bypassing labour intensive requirement and biases of laser capture microdissection.
Conclusion: These results provide the initial evidence of improved and cost-efficient analytics for the investigation of cancer cell microenvironment, using species-specificity arrays specifically designed for xenografts models.
Figures


References
-
- Finak G, Sadekova S, Pepin F, Hallett M, Meterissian S, Halwani F, Khetani K, Souleimanova M, Zabolotny B, Omeroglu A, Park M. Gene expression signatures of morphologically normal breast tissue identify basal-like tumors. Breast cancer research: BCR. 2006;7(5):R58. doi: 10.1186/bcr1608. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

