Integrative analysis reveals disease-associated genes and biomarkers for prostate cancer progression
- PMID: 25080090
- PMCID: PMC4110715
- DOI: 10.1186/1755-8794-7-S1-S3
Integrative analysis reveals disease-associated genes and biomarkers for prostate cancer progression
Abstract
Background: Prostate cancer is one of the most common complex diseases with high leading cause of death in men. Identifications of prostate cancer associated genes and biomarkers are thus essential as they can gain insights into the mechanisms underlying disease progression and advancing for early diagnosis and developing effective therapies.
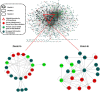
Methods: In this study, we presented an integrative analysis of gene expression profiling and protein interaction network at a systematic level to reveal candidate disease-associated genes and biomarkers for prostate cancer progression. At first, we reconstructed the human prostate cancer protein-protein interaction network (HPC-PPIN) and the network was then integrated with the prostate cancer gene expression data to identify modules related to different phases in prostate cancer. At last, the candidate module biomarkers were validated by its predictive ability of prostate cancer progression.
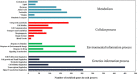
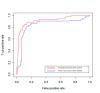
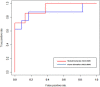
Results: Different phases-specific modules were identified for prostate cancer. Among these modules, transcription Androgen Receptor (AR) nuclear signaling and Epidermal Growth Factor Receptor (EGFR) signalling pathway were shown to be the pathway targets for prostate cancer progression. The identified candidate disease-associated genes showed better predictive ability of prostate cancer progression than those of published biomarkers. In context of functional enrichment analysis, interestingly candidate disease-associated genes were enriched in the nucleus and different functions were encoded for potential transcription factors, for examples key players as AR, Myc, ESR1 and hidden player as Sp1 which was considered as a potential novel biomarker for prostate cancer.
Conclusions: The successful results on prostate cancer samples demonstrated that the integrative analysis is powerful and useful approach to detect candidate disease-associate genes and modules which can be used as the potential biomarkers for prostate cancer progression. The data, tools and supplementary files for this integrative analysis are deposited at http://www.ibio-cn.org/HPC-PPIN/.
Figures




References
-
- Tricoli JV, Schoenfeldt M, Conley BA. Detection of prostate cancer and predicting progression: current and future diagnostic markers. Clin Cancer Res. 2004;7(12 Pt 1):3943–3953. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

