Computational models of liver fibrosis progression for hepatitis C virus chronic infection
- PMID: 25081062
- PMCID: PMC4120150
- DOI: 10.1186/1471-2105-15-S8-S5
Computational models of liver fibrosis progression for hepatitis C virus chronic infection
Abstract
Background: Chronic infection with hepatitis C virus (HCV) is a risk factor for liver diseases such as fibrosis, cirrhosis and hepatocellular carcinoma. HCV genetic heterogeneity was hypothesized to be associated with severity of liver disease. However, no reliable viral markers predicting disease severity have been identified. Here, we report the utility of sequences from 3 HCV 1b genomic regions, Core, NS3 and NS5b, to identify viral genetic markers associated with fast and slow rate of fibrosis progression (RFP) among patients with and without liver transplantation (n = 42).
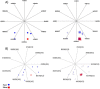
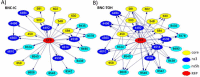
Methods: A correlation-based feature selection (CFS) method was used to detect and identify RFP-relevant viral markers. Machine-learning techniques, linear projection (LP) and Bayesian Networks (BN), were used to assess and identify associations between the HCV sequences and RFP.
Results: Both clustering of HCV sequences in LP graphs using physicochemical properties of nucleotides and BN analysis using polymorphic sites showed similarities among HCV variants sampled from patients with a similar RFP, while distinct HCV genetic properties were found associated with fast or slow RFP. Several RFP-relevant HCV sites were identified. Computational models parameterized using the identified sites accurately associated HCV strains with RFP in 70/30 split cross-validation (90-95% accuracy) and in validation tests (85-90% accuracy). Validation tests of the models constructed for patients with or without liver transplantation suggest that the RFP-relevant genetic markers identified in the HCV Core, NS3 and NS5b genomic regions may be useful for the prediction of RFP regardless of transplant status of patients.
Conclusions: The apparent strong genetic association to RFP suggests that HCV genetic heterogeneity has a quantifiable effect on severity of liver disease, thus presenting opportunity for developing genetic assays for measuring virulence of HCV strains in clinical and public health settings.
Figures

References
-
- Adam R, Karam V, Delvart V, O'Grady J, Mirza D, Klempnauer J, Castaing D, Neuhaus P, Jamieson N, Salizzoni M. et al.Evolution of indications and results of liver transplantation in Europe. A report from the European Liver Transplant Registry (ELTR) J Hepatol. 2012;57:675–688. doi: 10.1016/j.jhep.2012.04.015. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

