The evolutionary genetics of highly divergent alleles of the mimicry locus in Papilio dardanus
- PMID: 25081189
- PMCID: PMC4262259
- DOI: 10.1186/1471-2148-14-140
The evolutionary genetics of highly divergent alleles of the mimicry locus in Papilio dardanus
Abstract
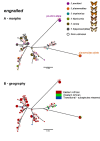
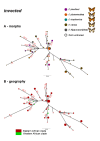
Background: The phylogenetic history of genes underlying phenotypic diversity can offer insight into the evolutionary origin of adaptive traits. This is especially true where single genes have large phenotypic effects, for example in determining polymorphic mimicry in butterflies. Here, we characterise the evolutionary history of two candidate genes for the mimicry switch in the polymorphic Batesian mimic Papilio dardanus coding for the transcription factors engrailed and invected.
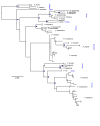
Results: We show that phased haplotypes associated with the dominant morphs f. poultoni and f. planemoides are phylogenetically highly divergent, in particular at non-synonymous sites. Some non-synonymous changes are shared between the divergent alleles suggesting either convergence or a shared ancestry. Gene trees for invected do not show this pattern. Despite their great divergence, all engrailed alleles of P. dardanus were monophyletic with respect to alleles of closely related species. Phylogenetic analyses therefore reveal no evidence for introgression from other species. A McDonald-Kreitman test conducted on a population sample from South Africa confirms a significant excess of intraspecific non-synonymous diversity in P. dardanus engrailed, suggesting long-term balanced polymorphism at this locus.
Conclusions: The divergence between engrailed haplotypes suggests an evolutionary history distorted by selection with multiple changes reflecting recurrent selective sweeps. The high level of intraspecific polymorphism observed is characteristic of balancing selection on this locus, as expected if the gene engrailed is under phenotypic selection for the maintenance of multiple mimetic morphs. Non-synonymous changes in key functional portions of a major transcription factor are likely to be deleterious but if maintained in a dominant allele at low frequency, heterozygosity would reduce the associated genetic load.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

