Tetra detector analysis of membrane proteins
- PMID: 25081744
- PMCID: PMC4178957
- DOI: 10.1002/0471140864.ps2910s77
Tetra detector analysis of membrane proteins
Abstract
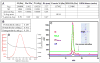
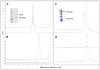
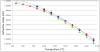
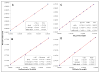
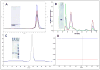
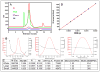
Well-characterized membrane protein detergent complexes (PDC) that are pure, homogenous, and stable, with minimized excess detergent micelles, are essential for functional assays and crystallization studies. Procedural steps to measure the mass, size, shape, homogeneity, and molecular composition of PDCs and their host detergent micelles using size-exclusion chromatography (SEC) with a Viscotek Tetra Detector Array (TDA; absorbance, refractive index, light scattering, and viscosity detectors) are presented in this unit. The value of starting with a quality PDC sample, the precision and accuracy of the results, and the use of a digital benchtop refractometer are emphasized. An alternate and simplified purification and characterization approach using SEC with dual absorbance and refractive index detectors to optimize detergent and lipid concentration while measuring the PDC homogeneity is also described. Applications relative to purification and characterization goals are illustrated as well.
Keywords: differential pressure viscometer; intrinsic viscosity; membrane proteins; refractive index; tetra detector array and analysis.
Copyright © 2014 John Wiley & Sons, Inc.
Figures










References
-
- Ball V, Ramsden JJ. Buffer dependence of refractive Index increments of protein solutions. Biopolymers. 1998;46:489–492.
-
- Barth HG, Boyes BE, Jackson C. Size exclusion chromatography. Analytical Chemistry. 1994;6(12):595R–620R. - PubMed
-
- Bohdanechy M, Kovar J. Viscosity of Polymer Solutions. Elsevier Scientific Publication; Amsterdam, Oxford, New York: 1982.
-
- Bohidar H. Light scattering and viscosity study of heat aggregation of insulin. Biopolymers. 1998;45:1–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

