Next-generation sequencing reveals large connected networks of intra-host HCV variants
- PMID: 25081811
- PMCID: PMC4120142
- DOI: 10.1186/1471-2164-15-S5-S4
Next-generation sequencing reveals large connected networks of intra-host HCV variants
Abstract
Background: Next-generation sequencing (NGS) allows for sampling numerous viral variants from infected patients. This provides a novel opportunity to represent and study the mutational landscape of Hepatitis C Virus (HCV) within a single host.
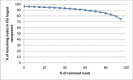
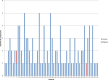
Results: Intra-host variants of the HCV E1/E2 region were extensively sampled from 58 chronically infected patients. After NGS error correction, the average number of reads and variants obtained from each sample were 3202 and 464, respectively. The distance between each pair of variants was calculated and networks were created for each patient, where each node is a variant and two nodes are connected by a link if the nucleotide distance between them is 1. The work focused on large components having > 5% of all reads, which in average account for 93.7% of all reads found in a patient.
Conclusions: Most intra-host variants are organized into distinct single-mutation components that are: well separated from each other, represent genetic distances between viral variants, robust to sampling, reproducible and likely seeded during transmission events. Facilitated by NGS, large components offer a novel evolutionary framework for genetic analysis of intra-host viral populations and understanding transmission, immune escape and drug resistance.
Figures



References
-
- Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: New estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2012. - PubMed
-
- Hsu CS, Liu CJ, Lai MY, Chen PJ, Kao JH, Chen DS. Early viral kinetics during treatment of chronic hepatitis C virus infection with pegylated interferon alpha plus ribavirin in Taiwan. Intervirology. 2007;15(4):310–315. - PubMed
-
- Aghemo A, Rumi MG, Colombo M. Pegylated IFN-alpha2a and ribavirin in the treatment of hepatitis C. Expert review of anti-infective therapy. 2009;15(8):925–935. - PubMed
-
- Simmonds P. Genetic diversity and evolution of hepatitis C virus - 15 years on. J Gen Virol. 2004;15:3173–3188. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

