H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex
- PMID: 25082344
- PMCID: PMC4139060
- DOI: 10.7554/eLife.02833
H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex
Abstract
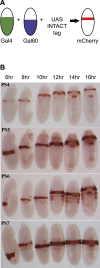
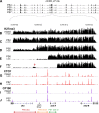
The bithorax complex (BX-C) in Drosophila melanogaster is a cluster of homeotic genes that determine body segment identity. Expression of these genes is governed by cis-regulatory domains, one for each parasegment. Stable repression of these domains depends on Polycomb Group (PcG) functions, which include trimethylation of lysine 27 of histone H3 (H3K27me3). To search for parasegment-specific signatures that reflect PcG function, chromatin from single parasegments was isolated and profiled. The H3K27me3 profiles across the BX-C in successive parasegments showed a 'stairstep' pattern that revealed sharp boundaries of the BX-C regulatory domains. Acetylated H3K27 was broadly enriched across active domains, in a pattern complementary to H3K27me3. The CCCTC-binding protein (CTCF) bound the borders between H3K27 modification domains; it was retained even in parasegments where adjacent domains lack H3K27me3. These findings provide a molecular definition of the homeotic domains, and implicate precisely positioned H3K27 modifications as a central determinant of segment identity.
Keywords: CTCF; Polycomb; bithorax complex; chromatin.
Copyright © 2014, Bowman et al.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures





References
-
- Bender W, Hudson A. 2000. P element homing to the Drosophila bithorax complex. Development 127:3981–3992 - PubMed
-
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, Ishikura T, Long HK, Sheahan TW, Brockdorff N, Kessler BM, Koseki H, Klose RJ. 2014. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157:1445–1459. doi: 10.1016/j.cell.2014.05.004 - DOI - PMC - PubMed
-
- Boulet AM, Lloyd A, Sakonju S. 1991. Molecular definition of the morphogenetic and regulatory functions and the cis-regulatory elements of the Drosophila Abd-B homeotic gene. Development 111:393–405 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

