Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia
- PMID: 25083870
- PMCID: PMC4131209
- DOI: 10.1016/j.cell.2014.05.049
Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia
Abstract
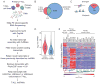
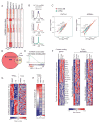
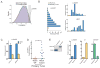
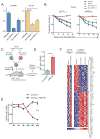
Notch signaling is a key developmental pathway that is subject to frequent genetic and epigenetic perturbations in many different human tumors. Here we investigate whether long noncoding RNA (lncRNA) genes, in addition to mRNAs, are key downstream targets of oncogenic Notch1 in human T cell acute lymphoblastic leukemia (T-ALL). By integrating transcriptome profiles with chromatin state maps, we have uncovered many previously unreported T-ALL-specific lncRNA genes, a fraction of which are directly controlled by the Notch1/Rpbjκ activator complex. Finally we have shown that one specific Notch-regulated lncRNA, LUNAR1, is required for efficient T-ALL growth in vitro and in vivo due to its ability to enhance IGF1R mRNA expression and sustain IGF1 signaling. These results confirm that lncRNAs are important downstream targets of the Notch signaling pathway, and additionally they are key regulators of the oncogenic state in T-ALL.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

References
-
- Boumil RM, Lee JT. Forty years of decoding the silence in X-chromosome inactivation. Hum Mol Genet. 2001;10:2225–2232. - PubMed
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- 5R01CA169784/CA/NCI NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- R01 CA133379/CA/NCI NIH HHS/United States
- 1R01GM088847/GM/NIGMS NIH HHS/United States
- R01 CA149655/CA/NCI NIH HHS/United States
- R01 CA105129/CA/NCI NIH HHS/United States
- 5R01CA173636/CA/NCI NIH HHS/United States
- 5T32GM0072-38/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- 5T32CA009161-37/CA/NCI NIH HHS/United States
- R01 CA169784/CA/NCI NIH HHS/United States
- U24 CA114766/CA/NCI NIH HHS/United States
- 1R01CA105129/CA/NCI NIH HHS/United States
- U10 CA98543/CA/NCI NIH HHS/United States
- R01 GM088847/GM/NIGMS NIH HHS/United States
- K99 CA188293/CA/NCI NIH HHS/United States
- R01 CA173636/CA/NCI NIH HHS/United States
- T32 CA009161/CA/NCI NIH HHS/United States
- 1R01CA133379/CA/NCI NIH HHS/United States
- 1R01CA149655/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

