Characterization of microbiota composition and presence of selected antibiotic resistance genes in carriage water of ornamental fish
- PMID: 25084116
- PMCID: PMC4118911
- DOI: 10.1371/journal.pone.0103865
Characterization of microbiota composition and presence of selected antibiotic resistance genes in carriage water of ornamental fish
Abstract
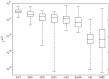
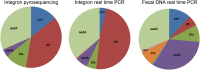
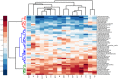
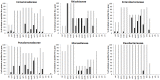
International trade with ornamental fish is gradually recognized as an important source of a wide range of different antibiotic resistant bacteria. In this study we therefore characterized the prevalence of selected antibiotic resistance genes in the microbiota found in the carriage water of ornamental fish originating from 3 different continents. Real-time PCR quantification showed that the sul1 gene was present in 11 out of 100 bacteria. tet(A) was present in 6 out of 100 bacteria and strA, tet(G), sul2 and aadA were present in 1-2 copies per 100 bacteria. Class I integrons were quite common in carriage water microbiota, however, pyrosequencing showed that only 12 different antibiotic gene cassettes were present in class I integrons. The microbiota characterized by pyrosequencing of the V3/V4 variable region of 16S rRNA genes consisted of Proteobacteria (48%), Bacteroidetes (29.5%), Firmicutes (17.8%), Actinobacteria (2.1%) and Fusobacteria (1.6%). Correlation analysis between antibiotic resistance gene prevalence and microbiota composition verified by bacterial culture showed that major reservoirs of sul1 sul2, tet(A), tet(B) tet(G), cat, cml, bla, strA, aacA, aph and aadA could be found among Alpha-, Beta- and Gammaproteobacteria with representatives of Enterobacteriaceae, Pseudomonadaceae, Rhizobiaceae and Comamonadaceae being those most positively associated with the tested antibiotic resistance genes.
Conflict of interest statement
Figures


References
-
- Cizek A, Dolejska M, Sochorova R, Strachotova K, Piackova V, et al. (2010) Antimicrobial resistance and its genetic determinants in aeromonads isolated in ornamental (koi) carp (Cyprinus carpio koi) and common carp (Cyprinus carpio). Vet Microbiol 142: 435–9. - PubMed
-
- Lim LC, Dhert P, Sorgeloos P (2003) Recent developments and improvements in ornamental fish packaging systems for air transport. Aquac Res 34: 923–35.
-
- Boyd D, Peters GA, Cloeckaert A, Boumedine KS, Chaslus-Dancla E, et al. (2001) Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar agona. J Bacteriol 183: 5725–32. - PMC - PubMed
-
- Hradecka H, Karasova D, Rychlik I (2008) Characterization of Salmonella enterica serovar Typhimurium conjugative plasmids transferring resistance to antibiotics and their interaction with the virulence plasmid. J Antimicrob Chemother 62: 938–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

