Sequence divergence and diversity suggests ongoing functional diversification of vertebrate NAD metabolism
- PMID: 25084685
- PMCID: PMC4248024
- DOI: 10.1016/j.dnarep.2014.07.005
Sequence divergence and diversity suggests ongoing functional diversification of vertebrate NAD metabolism
Abstract
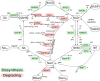
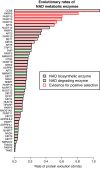
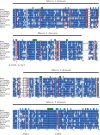
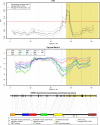
NAD is not only an important cofactor in redox reactions but has also received attention in recent years because of its physiological importance in metabolic regulation, DNA repair and signaling. In contrast to the redox reactions, these regulatory processes involve degradation of NAD and therefore necessitate a constant replenishment of its cellular pool. NAD biosynthetic enzymes are common to almost all species in all clades, but the number of NAD degrading enzymes varies substantially across taxa. In particular, vertebrates, including humans, have a manifold of NAD degrading enzymes which require a high turnover of NAD. As there is currently a lack of a systematic study of how natural selection has shaped enzymes involved in NAD metabolism we conducted a comprehensive evolutionary analysis based on intraspecific variation and interspecific divergence. We compare NAD biosynthetic and degrading enzymes in four eukaryotic model species and subsequently focus on human NAD metabolic enzymes and their orthologs in other vertebrates. We find that the majority of enzymes involved in NAD metabolism are subject to varying levels of purifying selection. While NAD biosynthetic enzymes appear to experience a rather high level of evolutionary constraint, there is evidence for positive selection among enzymes mediating NAD-dependent signaling. This is particularly evident for members of the PARP family, a diverse protein family involved in DNA damage repair and programmed cell death. Based on haplotype information and substitution rate analysis we pinpoint sites that are potential targets of positive selection. We also link our findings to a three dimensional structure, which suggests that positive selection occurs in domains responsible for DNA binding and polymerization rather than the NAD catalytic domain. Taken together, our results indicate that vertebrate NAD metabolism is still undergoing functional diversification.
Keywords: NAD metabolism; PARP; Pathway evolution; Positive selection.
Crown Copyright © 2014. Published by Elsevier B.V. All rights reserved.
Figures
Similar articles
-
Molecular evolutionary patterns of NAD+/Sirtuin aging signaling pathway across taxa.PLoS One. 2017 Aug 2;12(8):e0182306. doi: 10.1371/journal.pone.0182306. eCollection 2017. PLoS One. 2017. PMID: 28767699 Free PMC article.
-
New Insights into the Roles of NAD+-Poly(ADP-ribose) Metabolism and Poly(ADP-ribose) Glycohydrolase.Curr Protein Pept Sci. 2016;17(7):668-682. doi: 10.2174/1389203717666160419150014. Curr Protein Pept Sci. 2016. PMID: 27817743 Review.
-
Identification of novel components of NAD-utilizing metabolic pathways and prediction of their biochemical functions.Mol Biosyst. 2012 Jun;8(6):1661-77. doi: 10.1039/c2mb05487f. Epub 2012 Mar 7. Mol Biosyst. 2012. PMID: 22399070
-
Interplay between compartmentalized NAD+ synthesis and consumption: a focus on the PARP family.Genes Dev. 2020 Mar 1;34(5-6):254-262. doi: 10.1101/gad.335109.119. Epub 2020 Feb 6. Genes Dev. 2020. PMID: 32029457 Free PMC article. Review.
-
New perspectives on the plant PARP family: Arabidopsis PARP3 is inactive, and PARP1 exhibits predominant poly (ADP-ribose) polymerase activity in response to DNA damage.BMC Plant Biol. 2019 Aug 19;19(1):364. doi: 10.1186/s12870-019-1958-9. BMC Plant Biol. 2019. PMID: 31426748 Free PMC article.
Cited by
-
Targeting Nuclear NAD+ Synthesis Inhibits DNA Repair, Impairs Metabolic Adaptation and Increases Chemosensitivity of U-2OS Osteosarcoma Cells.Cancers (Basel). 2020 May 7;12(5):1180. doi: 10.3390/cancers12051180. Cancers (Basel). 2020. PMID: 32392755 Free PMC article.
-
Molecular evolutionary patterns of NAD+/Sirtuin aging signaling pathway across taxa.PLoS One. 2017 Aug 2;12(8):e0182306. doi: 10.1371/journal.pone.0182306. eCollection 2017. PLoS One. 2017. PMID: 28767699 Free PMC article.
-
Nicotinamide N-Methyltransferase in Health and Cancer.Int J Tryptophan Res. 2017 Jun 30;10:1178646917691739. doi: 10.1177/1178646917691739. eCollection 2017. Int J Tryptophan Res. 2017. PMID: 35185340 Free PMC article. Review.
-
Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.Open Biol. 2020 Nov;10(11):200237. doi: 10.1098/rsob.200237. Epub 2020 Nov 18. Open Biol. 2020. PMID: 33202171 Free PMC article.
-
Identification of evolutionary and kinetic drivers of NAD-dependent signaling.Proc Natl Acad Sci U S A. 2019 Aug 6;116(32):15957-15966. doi: 10.1073/pnas.1902346116. Epub 2019 Jul 24. Proc Natl Acad Sci U S A. 2019. PMID: 31341085 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

