Integrated genome-wide association, coexpression network, and expression single nucleotide polymorphism analysis identifies novel pathway in allergic rhinitis
- PMID: 25085501
- PMCID: PMC4127082
- DOI: 10.1186/1755-8794-7-48
Integrated genome-wide association, coexpression network, and expression single nucleotide polymorphism analysis identifies novel pathway in allergic rhinitis
Abstract
Background: Allergic rhinitis is a common disease whose genetic basis is incompletely explained. We report an integrated genomic analysis of allergic rhinitis.
Methods: We performed genome wide association studies (GWAS) of allergic rhinitis in 5633 ethnically diverse North American subjects. Next, we profiled gene expression in disease-relevant tissue (peripheral blood CD4+ lymphocytes) collected from subjects who had been genotyped. We then integrated the GWAS and gene expression data using expression single nucleotide (eSNP), coexpression network, and pathway approaches to identify the biologic relevance of our GWAS.
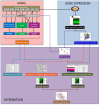
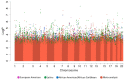
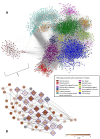
Results: GWAS revealed ethnicity-specific findings, with 4 genome-wide significant loci among Latinos and 1 genome-wide significant locus in the GWAS meta-analysis across ethnic groups. To identify biologic context for these results, we constructed a coexpression network to define modules of genes with similar patterns of CD4+ gene expression (coexpression modules) that could serve as constructs of broader gene expression. 6 of the 22 GWAS loci with P-value ≤ 1x10-6 tagged one particular coexpression module (4.0-fold enrichment, P-value 0.0029), and this module also had the greatest enrichment (3.4-fold enrichment, P-value 2.6 × 10-24) for allergic rhinitis-associated eSNPs (genetic variants associated with both gene expression and allergic rhinitis). The integrated GWAS, coexpression network, and eSNP results therefore supported this coexpression module as an allergic rhinitis module. Pathway analysis revealed that the module was enriched for mitochondrial pathways (8.6-fold enrichment, P-value 4.5 × 10-72).
Conclusions: Our results highlight mitochondrial pathways as a target for further investigation of allergic rhinitis mechanism and treatment. Our integrated approach can be applied to provide biologic context for GWAS of other diseases.
Figures


Similar articles
-
The association between sixteen genome-wide association studies-related allergic diseases loci and childhood allergic rhinitis in a Chinese Han population.Cytokine. 2018 Nov;111:162-170. doi: 10.1016/j.cyto.2018.08.022. Epub 2018 Aug 28. Cytokine. 2018. PMID: 30170133
-
Functional variants of 17q12-21 are associated with allergic asthma but not allergic rhinitis.J Allergy Clin Immunol. 2016 Mar;137(3):758-66.e3. doi: 10.1016/j.jaci.2015.08.038. Epub 2015 Oct 23. J Allergy Clin Immunol. 2016. PMID: 26483175
-
A conserved BDNF, glutamate- and GABA-enriched gene module related to human depression identified by coexpression meta-analysis and DNA variant genome-wide association studies.PLoS One. 2014 Mar 7;9(3):e90980. doi: 10.1371/journal.pone.0090980. eCollection 2014. PLoS One. 2014. PMID: 24608543 Free PMC article.
-
Genetics and Epigenetics in Allergic Rhinitis.Genes (Basel). 2021 Dec 17;12(12):2004. doi: 10.3390/genes12122004. Genes (Basel). 2021. PMID: 34946955 Free PMC article. Review.
-
Genetics of allergic diseases.Immunol Allergy Clin North Am. 2015 Feb;35(1):19-44. doi: 10.1016/j.iac.2014.09.014. Epub 2014 Nov 21. Immunol Allergy Clin North Am. 2015. PMID: 25459575 Free PMC article. Review.
Cited by
-
Resolving the etiology of atopic disorders by using genetic analysis of racial ancestry.J Allergy Clin Immunol. 2016 Sep;138(3):676-699. doi: 10.1016/j.jaci.2016.02.045. Epub 2016 Jun 11. J Allergy Clin Immunol. 2016. PMID: 27297995 Free PMC article. Review.
-
Replication study of susceptibility variants associated with allergic rhinitis and allergy in Han Chinese.Allergy Asthma Clin Immunol. 2020 Feb 11;16:13. doi: 10.1186/s13223-020-0411-9. eCollection 2020. Allergy Asthma Clin Immunol. 2020. PMID: 32082391 Free PMC article.
-
Systems biology and in vitro validation identifies family with sequence similarity 129 member A (FAM129A) as an asthma steroid response modulator.J Allergy Clin Immunol. 2018 Nov;142(5):1479-1488.e12. doi: 10.1016/j.jaci.2017.11.059. Epub 2018 Mar 2. J Allergy Clin Immunol. 2018. PMID: 29410046 Free PMC article. Clinical Trial.
-
International Consensus Statement on Allergy and Rhinology: Allergic Rhinitis.Int Forum Allergy Rhinol. 2018 Feb;8(2):108-352. doi: 10.1002/alr.22073. Int Forum Allergy Rhinol. 2018. PMID: 29438602 Free PMC article.
-
Combining omics data to identify genes associated with allergic rhinitis.Clin Epigenetics. 2017 Jan 18;9:3. doi: 10.1186/s13148-017-0310-1. eCollection 2017. Clin Epigenetics. 2017. PMID: 28149331 Free PMC article.
References
-
- Wallace DV, Dykewicz MS, Bernstein DI, Blessing-Moore J, Cox L, Khan DA, Lang DM, Nicklas RA, Oppenheimer J, Portnoy JM, Randolph CC, Schuller D, Spector SL, Tilles SA. The diagnosis and management of rhinitis: an updated practice parameter. J Allergy Clin Immunol. 2008;7:S1–S84. - PubMed
-
- Feijen M, Gerritsen J, Postma DS. Genetics of allergic disease. Br Med Bull. 2000;7:894–907. - PubMed
-
- Davila I, Mjullol J, Ferrer M, Bartra J, del Cuvillo A, Montoro J, Jauregui I, Sastre J, Valero A. Genetic aspects of allergic rhinitis. J Investig Allergol Clin Immunol. 2009;7:25–31. - PubMed
-
- Ramasamy A, Curjuric I, Coin LJ, Kumar A, McArdle WL, Imboden M, Leynaert B, Kogevinas M, Schmid-Grendelmeier P, Pekkanen J, Wjst M, Bircher AJ, Sovio U, Rochat T, Hartikainen AL, Balding DJ, Jarvelin MR, Probst-Hensch N, Strachan DP, Jarvis DL. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol. 2011;7:996–1005. - PubMed
-
- Hindorff LA MJEBI, Morales J, Junkins HA, Hall PN, Klemm AK, Manolio TA, (European Bioinformatics Institute) A Catalog of Published Genome-Wide Association Studies. Available at: http://www.genome.gov/gwastudies Accessed September 19, 2013 2013.
Publication types
MeSH terms
Grants and funding
- AI077439/AI/NIAID NIH HHS/United States
- ES015794/ES/NIEHS NIH HHS/United States
- HL069167/HL/NHLBI NIH HHS/United States
- ES009581/ES/NIEHS NIH HHS/United States
- ES011627/ES/NIEHS NIH HHS/United States
- HL078885/HL/NHLBI NIH HHS/United States
- RD831861/RD/ORD VA/United States
- AI070503/AI/NIAID NIH HHS/United States
- HL079055/HL/NHLBI NIH HHS/United States
- HL101543/HL/NHLBI NIH HHS/United States
- 001/WHO_/World Health Organization/International
- HL101651/HL/NHLBI NIH HHS/United States
- R00 HL105663/HL/NHLBI NIH HHS/United States
- Z01 ES049019/ES/NIEHS NIH HHS/United States
- HL65899/HL/NHLBI NIH HHS/United States
- P30 ES007048/ES/NIEHS NIH HHS/United States
- ES007048/ES/NIEHS NIH HHS/United States
- HL083069/HL/NHLBI NIH HHS/United States
- K08 AI093538/AI/NIAID NIH HHS/United States
- HL085197/HL/NHLBI NIH HHS/United States
- U01 HL065899/HL/NHLBI NIH HHS/United States
- HL072414/HL/NHLBI NIH HHS/United States
- ES020801/ES/NIEHS NIH HHS/United States
- R37 HL066289/HL/NHLBI NIH HHS/United States
- P01ES011627/ES/NIEHS NIH HHS/United States
- HL076647/HL/NHLBI NIH HHS/United States
- HL087680/HL/NHLBI NIH HHS/United States
- AI061774/AI/NIAID NIH HHS/United States
- HL087699/HL/NHLBI NIH HHS/United States
- P30ES007048/ES/NIEHS NIH HHS/United States
- HL075419/HL/NHLBI NIH HHS/United States
- HL064307/HL/NHLBI NIH HHS/United States
- HL49596/HL/NHLBI NIH HHS/United States
- HL064313/HL/NHLBI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- K08AI093538/AI/NIAID NIH HHS/United States
- HL066289/HL/NHLBI NIH HHS/United States
- DK064695/DK/NIDDK NIH HHS/United States
- ES022719/ES/NIEHS NIH HHS/United States
- AI079139/AI/NIAID NIH HHS/United States
- R01 HL086601/HL/NHLBI NIH HHS/United States
- HL061768/HL/NHLBI NIH HHS/United States
- HL088133/HL/NHLBI NIH HHS/United States
- HL087665/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

