The Amyloid Precursor Protein is rapidly transported from the Golgi apparatus to the lysosome and where it is processed into beta-amyloid
- PMID: 25085554
- PMCID: PMC4237969
- DOI: 10.1186/s13041-014-0054-1
The Amyloid Precursor Protein is rapidly transported from the Golgi apparatus to the lysosome and where it is processed into beta-amyloid
Abstract
Background: Alzheimer's disease (AD) is characterized by cerebral deposition of β-amyloid peptide (Aβ). Aβ is produced by sequential cleavage of the Amyloid Precursor Protein (APP) by β- and γ-secretases. Many studies have demonstrated that the internalization of APP from the cell surface can regulate Aβ production, although the exact organelle in which Aβ is produced remains contentious. A number of recent studies suggest that intracellular trafficking also plays a role in regulating Aβ production, but these pathways are relatively under-studied. The goal of this study was to elucidate the intracellular trafficking of APP, and to examine the site of intracellular APP processing.
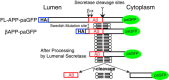
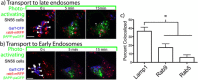
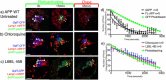
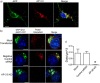
Results: We have tagged APP on its C-terminal cytoplasmic tail with photoactivatable Green Fluorescent Protein (paGFP). By photoactivating APP-paGFP in the Golgi, using the Golgi marker Galactosyltranferase fused to Cyan Fluorescent Protein (GalT-CFP) as a target, we are able to follow a population of nascent APP molecules from the Golgi to downstream compartments identified with compartment markers tagged with red fluorescent protein (mRFP or mCherry); including rab5 (early endosomes) rab9 (late endosomes) and LAMP1 (lysosomes). Because γ-cleavage of APP releases the cytoplasmic tail of APP including the photoactivated GFP, resulting in loss of fluorescence, we are able to visualize the cleavage of APP in these compartments. Using APP-paGFP, we show that APP is rapidly trafficked from the Golgi apparatus to the lysosome; where it is rapidly cleared. Chloroquine and the highly selective γ-secretase inhibitor, L685, 458, cause the accumulation of APP in lysosomes implying that APP is being cleaved by secretases in the lysosome. The Swedish mutation dramatically increases the rate of lysosomal APP processing, which is also inhibited by chloroquine and L685, 458. By knocking down adaptor protein 3 (AP-3; a heterotetrameric protein complex required for trafficking many proteins to the lysosome) using siRNA, we are able to reduce this lysosomal transport. Blocking lysosomal transport of APP reduces Aβ production by more than a third.
Conclusion: These data suggests that AP-3 mediates rapid delivery of APP to lysosomes, and that the lysosome is a likely site of Aβ production.
Figures




References
-
- Mebane-Sims I. Alzheimer's disease facts and figures. Alzheimers Dement. 2009;2009(5):234–270. - PubMed
-
- Hussain I, Powell D, Howlett DR, Tew DG, Meek TD, Chapman C, Gloger IS, Murphy KE, Southan CD, Ryan DM, Smith TS, Simmons DL, Walsh FS, Dingwall C, Christie G. Identification of a Novel Aspartic Protease (Asp 2) as β-Secretase. Mol Cell Neurosci. 1999;14:419–427. - PubMed
-
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

