Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer
- PMID: 25086042
- PMCID: PMC4176249
- DOI: 10.1074/jbc.M114.553818
Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer
Erratum in
- J Biol Chem. 2014 Dec 5;289(49):33877. Hessein, Mohamed [Corrected to Hassona, Mohamed D]
-
Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer.J Biol Chem. 2015 Oct 23;290(43):25850. doi: 10.1074/jbc.A115.553818. J Biol Chem. 2015. PMID: 26500294 Free PMC article. No abstract available.
-
Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer.J Biol Chem. 2017 Mar 10;292(10):4359. doi: 10.1074/jbc.A117.553818. J Biol Chem. 2017. PMID: 28283588 Free PMC article. No abstract available.
Abstract
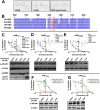
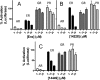
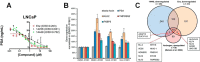
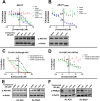
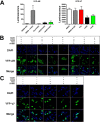
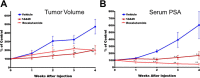
The androgen receptor (AR) is a transcription factor that has a pivotal role in the occurrence and progression of prostate cancer. The AR is activated by androgens that bind to its ligand-binding domain (LBD), causing the transcription factor to enter the nucleus and interact with genes via its conserved DNA-binding domain (DBD). Treatment for prostate cancer involves reducing androgen production or using anti-androgen drugs to block the interaction of hormones with the AR-LBD. Eventually the disease changes into a castration-resistant form of PCa where LBD mutations render anti-androgens ineffective or where constitutively active AR splice variants, lacking the LBD, become overexpressed. Recently, we identified a surfaced exposed pocket on the AR-DBD as an alternative drug-target site for AR inhibition. Here, we demonstrate that small molecules designed to selectively bind the pocket effectively block transcriptional activity of full-length and splice variant AR forms at low to sub-micromolar concentrations. The inhibition is lost when residues involved in drug interactions are mutated. Furthermore, the compounds did not impede nuclear localization of the AR and blocked interactions with chromatin, indicating the interference of DNA binding with the nuclear form of the transcription factor. Finally, we demonstrate the inhibition of gene expression and tumor volume in mouse xenografts. Our results indicate that the AR-DBD has a surface site that can be targeted to inhibit all forms of the AR, including enzalutamide-resistant and constitutively active splice variants and thus may serve as a potential avenue for the treatment of recurrent and metastatic prostate cancer.
Keywords: Androgen Receptor; DNA-binding Protein; Drug Discovery; Nuclear Receptor; Prostate Cancer.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Cutress M. L., Whitaker H. C., Mills I. G., Stewart M., Neal D. E. (2008) Structural basis for the nuclear import of the human androgen receptor. J. Cell Sci. 121, 957–968 - PubMed
-
- Lamont K. R., Tindall D. J. (2010) Androgen regulation of gene expression. Adv. Cancer Res. 107, 137–162 - PubMed
-
- Saad F., Miller K. (2014) Treatment options in castration-resistant prostate cancer: current therapies and emerging docetaxel-based regimens. Urol. Oncol. 32, 70–79 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

