Nrbf2 protein suppresses autophagy by modulating Atg14L protein-containing Beclin 1-Vps34 complex architecture and reducing intracellular phosphatidylinositol-3 phosphate levels
- PMID: 25086043
- PMCID: PMC4176228
- DOI: 10.1074/jbc.M114.561134
Nrbf2 protein suppresses autophagy by modulating Atg14L protein-containing Beclin 1-Vps34 complex architecture and reducing intracellular phosphatidylinositol-3 phosphate levels
Abstract
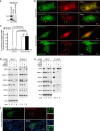
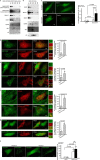
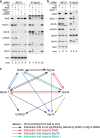
Autophagy is a tightly regulated lysosomal degradation pathway for maintaining cellular homeostasis and responding to stresses. Beclin 1 and its interacting proteins, including the class III phosphatidylinositol-3 kinase Vps34, play crucial roles in autophagy regulation in mammals. We identified nuclear receptor binding factor 2 (Nrbf2) as a Beclin 1-interacting protein from Becn1(-/-);Becn1-EGFP/+ mouse liver and brain. We also found that Nrbf2-Beclin 1 interaction required the N terminus of Nrbf2. We next used the human retinal pigment epithelial cell line RPE-1 as a model system and showed that transiently knocking down Nrbf2 by siRNA increased autophagic flux under both nutrient-rich and starvation conditions. To investigate the mechanism by which Nrbf2 regulates autophagy, we demonstrated that Nrbf2 interacted and colocalized with Atg14L, suggesting that Nrbf2 is a component of the Atg14L-containing Beclin 1-Vps34 complex. Moreover, ectopically expressed Nrbf2 formed cytosolic puncta that were positive for isolation membrane markers. These results suggest that Nrbf2 is involved in autophagosome biogenesis. Furthermore, we showed that Nrbf2 deficiency led to increased intracellular phosphatidylinositol-3 phosphate levels and diminished Atg14L-Vps34/Vps15 interactions, suggesting that Nrbf2-mediated Atg14L-Vps34/Vps15 interactions likely inhibit Vps34 activity. Therefore, we propose that Nrbf2 may interact with the Atg14L-containing Beclin 1-Vps34 protein complex to modulate protein-protein interactions within the complex, leading to suppression of Vps34 activity, autophagosome biogenesis, and autophagic flux. This work reveals a novel aspect of the intricate mechanism for the Beclin 1-Vps34 protein-protein interaction network to achieve precise control of autophagy.
Keywords: Autophagosome Biogenesis; Autophagy; Beclin 1; Cellular Regulation; Nrbf2; Protein Degradation; Protein-Protein Interaction; Subcellular Organelle; Vps34.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Suzuki K., Ohsumi Y. (2007) Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 581, 2156–2161 - PubMed
-
- Liu Y., Shoji-Kawata S., Sumpter R. M., Jr., Wei Y., Ginet V., Zhang L., Posner B., Tran K. A., Green D. R., Xavier R. J., Shaw S. Y., Clarke P. G., Puyal J., Levine B. (2013) Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc. Natl. Acad. Sci. U.S.A. 110, 20364–20371 - PMC - PubMed
-
- Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

