A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae
- PMID: 25086048
- PMCID: PMC4176211
- DOI: 10.1074/jbc.M114.593996
A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae
Abstract
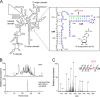
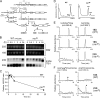
Biogenesis of eukaryotic ribosome is a complex event involving a number of non-ribosomal factors. During assembly of the ribosome, rRNAs are post-transcriptionally modified by 2'-O-methylation, pseudouridylation, and several base-specific modifications, which are collectively involved in fine-tuning translational fidelity and/or modulating ribosome assembly. By mass-spectrometric analysis, we demonstrated that N(4)-acetylcytidine (ac(4)C) is present at position 1773 in the 18 S rRNA of Saccharomyces cerevisiae. In addition, we found an essential gene, KRE33 (human homolog, NAT10), that we renamed RRA1 (ribosomal RNA cytidine acetyltransferase 1) encoding an RNA acetyltransferase responsible for ac(4)C1773 formation. Using recombinant Rra1p, we could successfully reconstitute ac(4)C1773 in a model rRNA fragment in the presence of both acetyl-CoA and ATP as substrates. Upon depletion of Rra1p, the 23 S precursor of 18 S rRNA was accumulated significantly, which resulted in complete loss of 18 S rRNA and small ribosomal subunit (40 S), suggesting that ac(4)C1773 formation catalyzed by Rra1p plays a critical role in processing of the 23 S precursor to yield 18 S rRNA. When nuclear acetyl-CoA was depleted by inactivation of acetyl-CoA synthetase 2 (ACS2), we observed temporal accumulation of the 23 S precursor, indicating that Rra1p modulates biogenesis of 40 S subunit by sensing nuclear acetyl-CoA concentration.
Keywords: Acetyl Coenzyme A (Acetyl-CoA); Acetyltransferase; RNA Modification; Ribosomal RNA Processing (rRNA Processing); Ribosome Assembly.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Warner J. R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 - PubMed
-
- Venema J., Tollervey D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33, 261–311 - PubMed
-
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. (2003) Ribosome assembly in eukaryotes. Gene 313, 17–42 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

